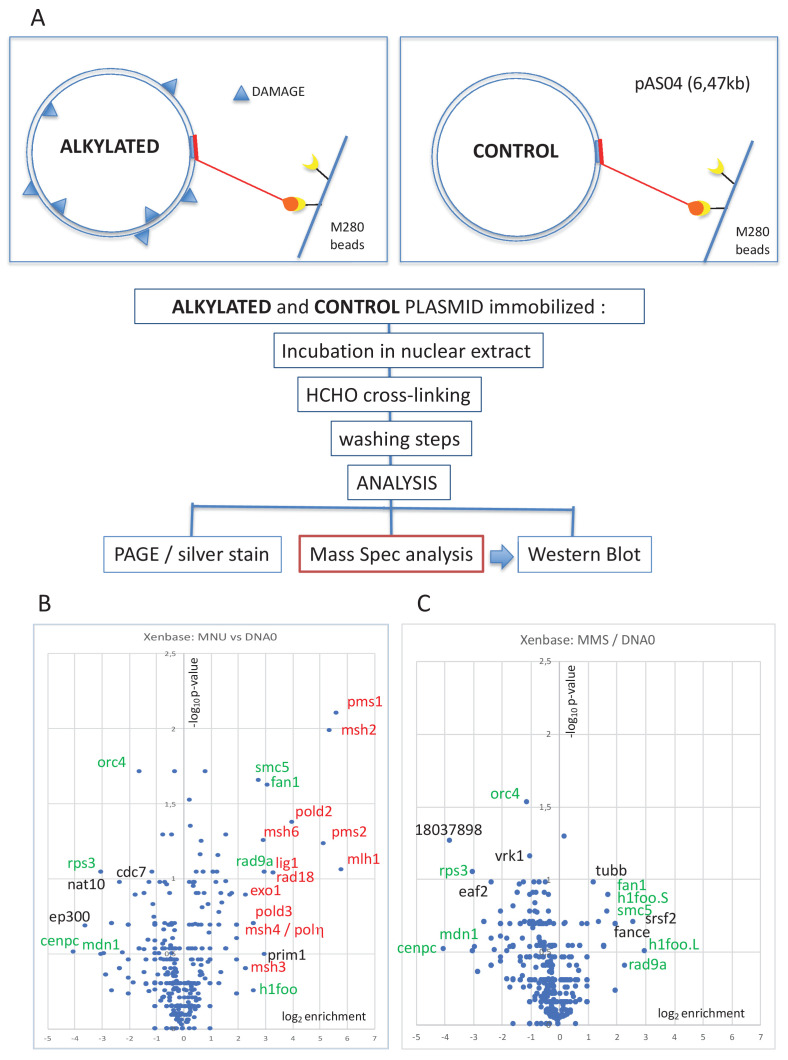
Figure 1. Pull-down of proteins that bind to alkylated versus untreated plasmid DNA.
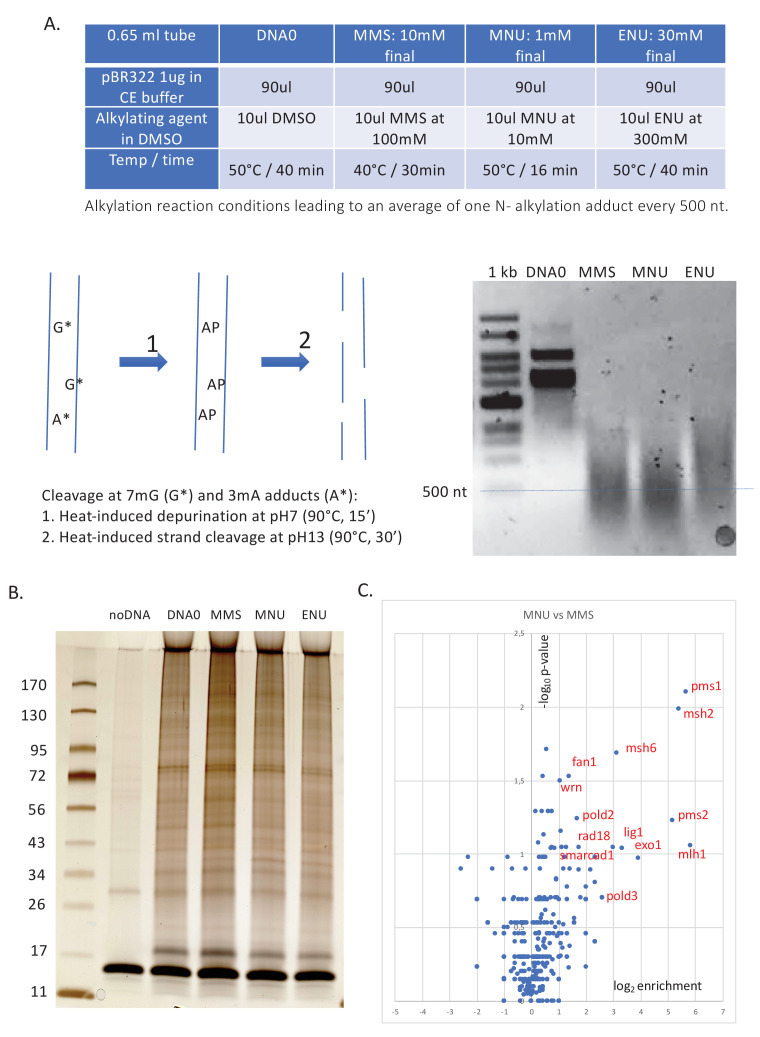
(A) Experimental workflow. Plasmid DNA (pAS04, 6.5 kb) was treated with alkylating agents under conditions leading to a similar extent of N-alkylation (≈ one alkaline cleavage site every 500 nt) (Figure 1—figure supplement 1A). Immobilized plasmid DNA was incubated in Xenopus nucleoplasmic extracts (NPE) for 10 min at room temperature under mild agitation. The reaction was stopped by addition of formaldehyde (0.8% final) to cross-link the protein-DNA complexes. The beads were processed and analyzed by polyacrylamide gel electrophoresis (PAGE) or by mass spectrometry (MS) as described in 'Materials and methods'. (B) Relative abundance of proteins captured on N-methyl-N-nitrosourea (MNU)-treated versus -untreated DNA0. Proteins captured on equal amounts of MNU-treated or -untreated plasmid were analyzed by label-free MS in triplicate. For all proteins, average spectral count values in the MNU-treated plasmid sample were divided by the average spectral count values in the DNA0 sample. The resulting ratio is plotted as its log2 value along x-axis. The statistical significance of the data is estimated by the p-value in the Student’s t-test and plotted as -log10p along y-axis. Proteins enriched on MNU versus untreated plasmid DNA appear on the right-side top corner and essentially turn out to be mismatch repair (MMR) proteins labeled in red (B). Data shown are analyzed using Xenbase database. (C) Relative abundance of proteins captured on methyl-methane sulfonate (MMS)-treated versus -untreated DNA0. Proteins captured on equal amounts of MMS-treated or -untreated plasmid were analyzed by label-free MS in triplicate. The data are analyzed and plotted as in panel (B) for MNU using Xenbase database. Proteins (labeled in green in B and C) are found enriched or excluded in both MMS versus DNA0 and MNU versus DNA0 plasmids. We suggest these proteins are recruited or excluded from binding to DNA by the abundant class of N-alkylation adducts that both MMS- and MNU-treated plasmids share in common (~27 N-alkyl adducts per plasmid).