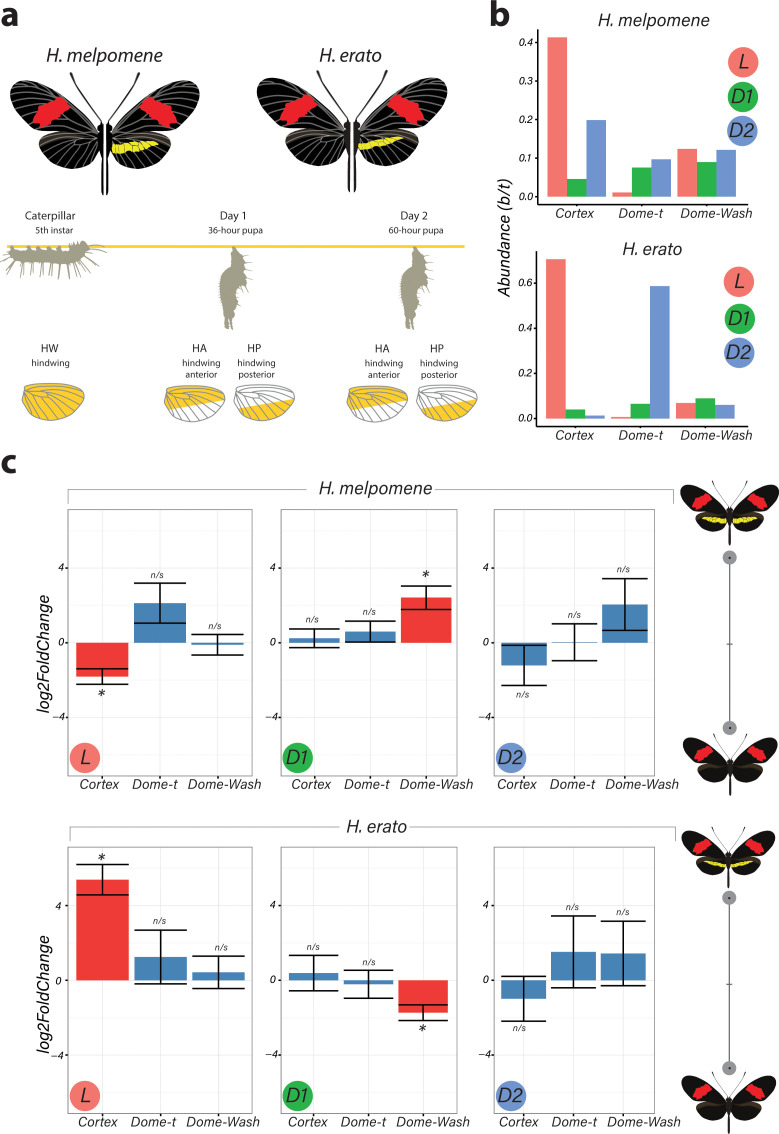
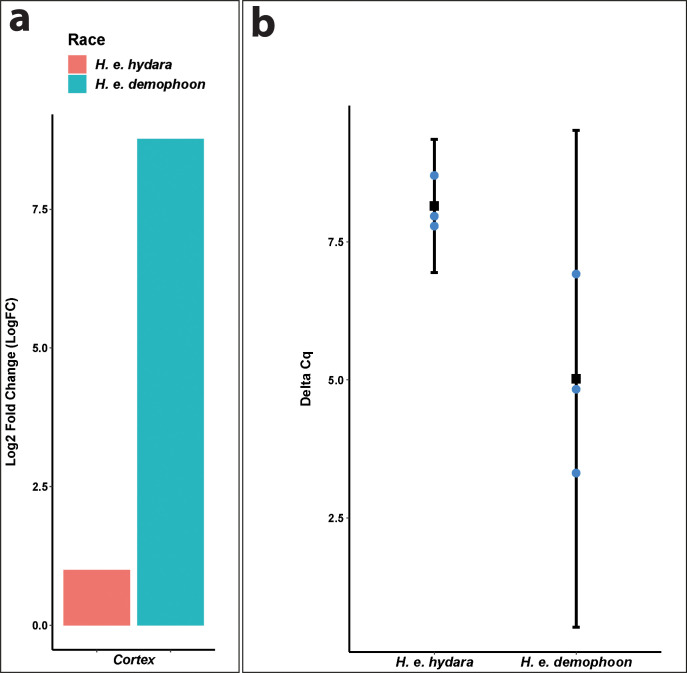
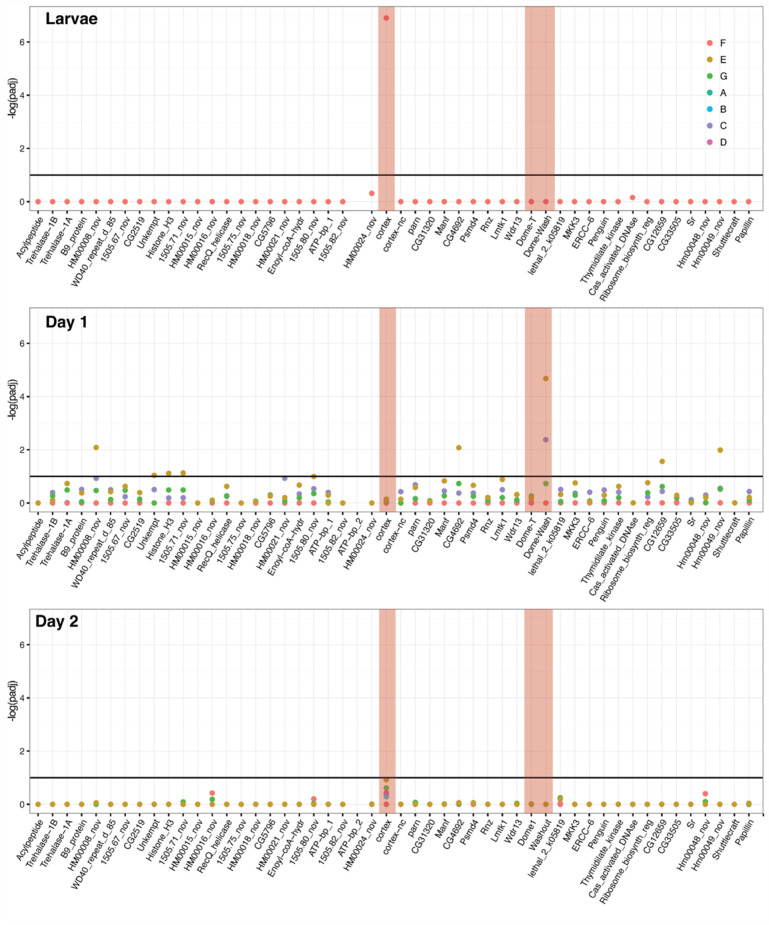
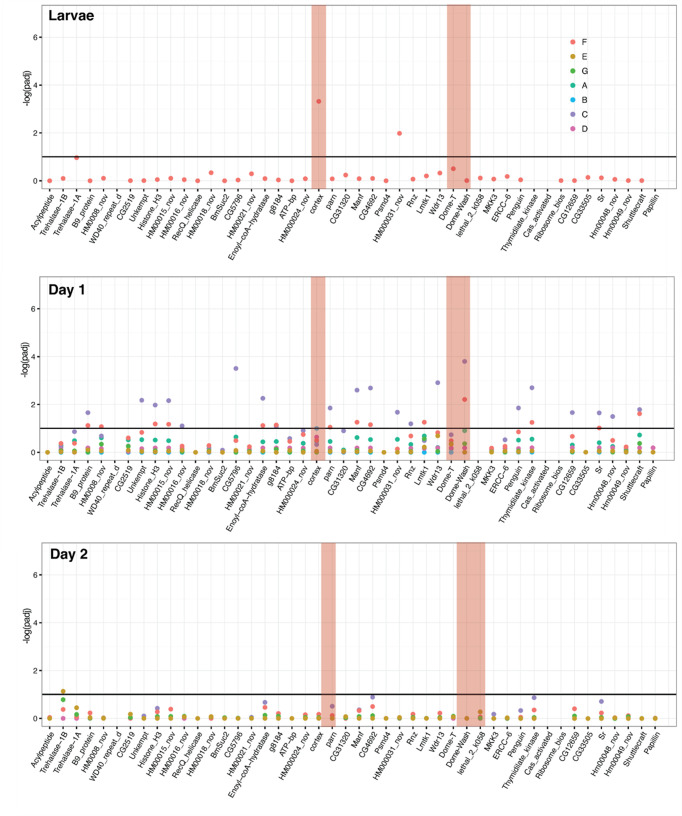
Figure 2. Differential expression of genes at Chromosome 15 implicate cortex as most likely candidate driving yellow bar differences.
(a) Hindwing tissue from co-mimetic morphs of H. melpomene and H. erato were collected at three developmental stages fifth-instar caterpillar, Day 1 Pupae (36hAPF) and Day 2 Pupae (60hAPF). For pupal tissue, hindwing tissue was dissected using the wing vein landmarks shown, corresponding to the future adult position of the hindwing yellow bar (dissection scheme based on Hanly et al., 2019). (b) Relative abundance of transcripts corresponding to the genes cortex, domeless-truncated, domeless/washout throughout developmental stages. (c) Log2FoldChange for the genes cortex, domeless-truncated (dome-t), domeless/washout (dome-wash) across developmental stages. Comparisons are for whole wing discs (Larvae, L) and across wing sections differing in the presence of a yellow bar in pupal wings (D1 and D2; see Figure 2—figure supplement 2: for depiction of contrasts analysed). *Adjusted p<0.05; n/s = not significant. N = 3 for each bar plot.