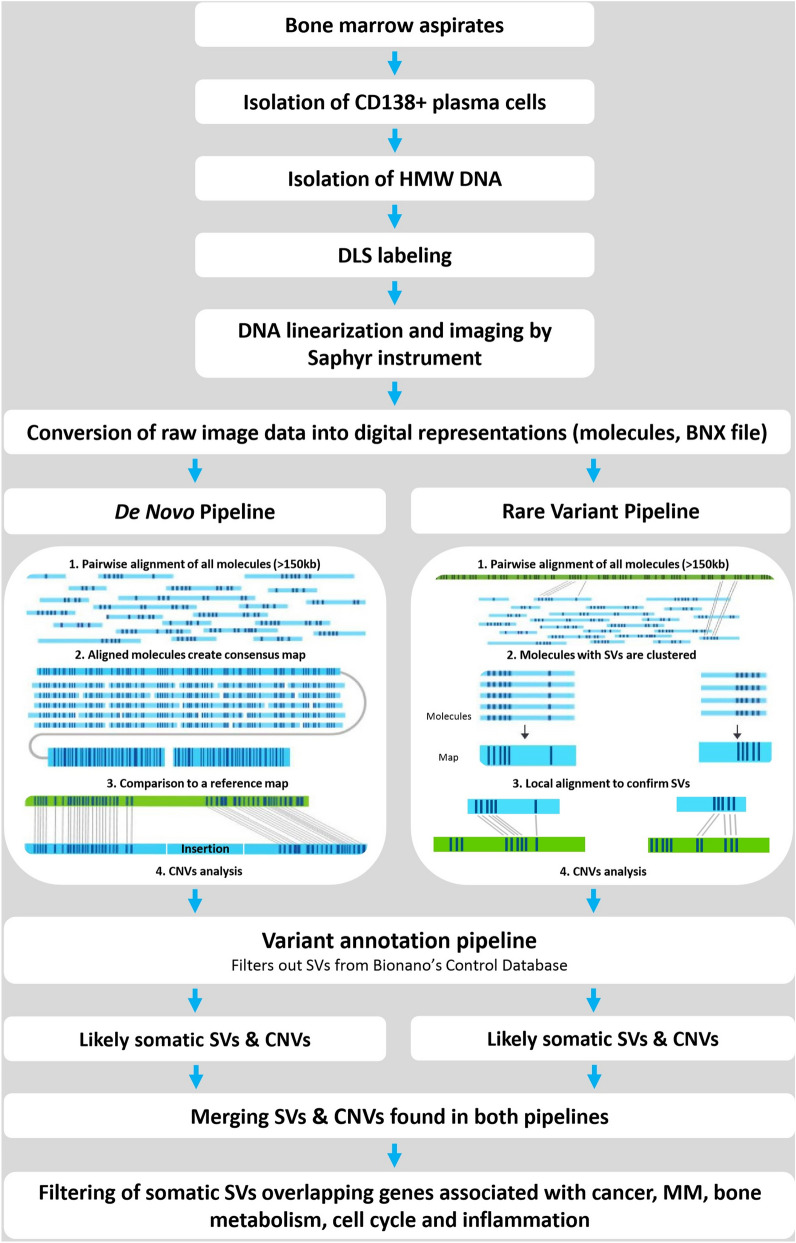
Figure 1.
Workflow of optical mapping and bioinformatics pipelines used. HMW DNA is isolated from CD138+ plasma cells of BM aspirates and labelled by DLS chemistry in specific sequences across entire genomes. Labelled DNA is loaded on the chip and linearised and visualised in a Saphyr instrument. Images are converted to BNX molecules. The architecture of the bioinformatics pipeline includes two pipelines (de novo and rare variant), constructing optical genome maps and comparing them with a human reference map (hg38), filtering detected variants for somatic SVs and merging data from both pipelines. The last step enables a comparison of the data with the gene panels created from NCBI gene datasets.