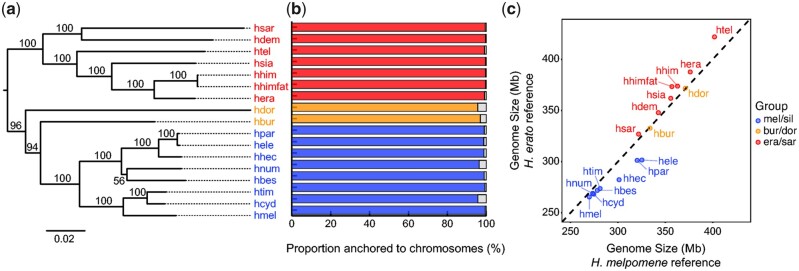
Fig. 1.
Reference-guided assemblies. (a) ML tree from whole mitochondrial genomes assembled here. Bootstrap values are shown next to the branches. The tree was rooted using the E. tales mitochondrial genome. (b) Proportion of the reference-scaffolded assemblies length anchored to chromosomes. The results are shown for the reference-guided assemblies mapped to the closest reference (either H. melpomene or H. erato demophoon; see supplementary table 2, Supplementary Material online). For a complete report of the results, see supplementary figure 37, Supplementary Material online. (c) Reference-scaffolded genome sizes using either H. melpomene (x axis) or H. erato demophoon (y axis) as the reference. The dashed line represents the expectation if there was a 1:1 correspondence. In all panels, subclade memberships are represented by different colors—melpomene/silvaniform (blue), burneyi + doris (yellow), erato/sara (red). Species codes for all the new reference-guided assemblies are as follows: hmel—H. melpomene; hcyd—H. cydno; htim—H. timareta; hbes—H. besckei; hnum—H. numata; hhec—H. hecale; hele—H. elevatus; hpar—H. pardalinus; hbur—H. burneyi; hdor—H. doris; hera—H. erato; hhimfat—H. himera; hhim—H. himera; hsia—H. hecalesia; htel—H. telesiphe; hdem—H. demeter; hsar—H. sara.