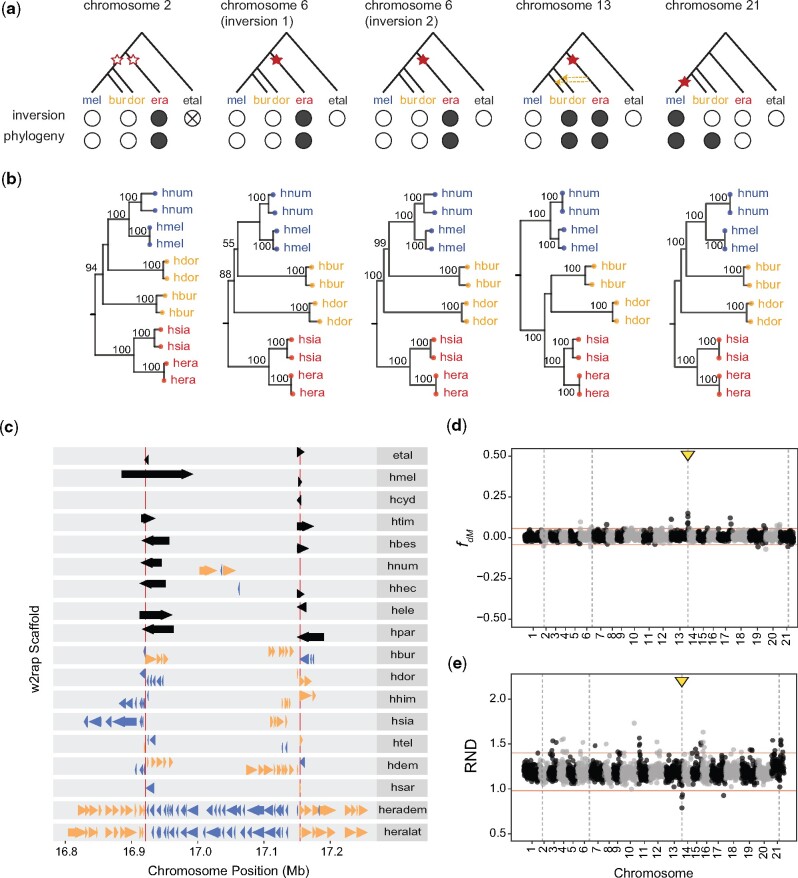
Fig. 4.
Large inversions fixed between the melpomene/silvaniform and erato/sara clades. (a) Possible scenario for the origin and sharing of the inversion. Stars represent inversions and the branch in which these likely took place. Empty stars are used when the inversion could have occurred in either of two branches. Introgression between branches is represented by arrows, the direction of the arrow indicating directionality. Full and empty circles represent the orientation of the inversion (empty circle with a cross indicates that the orientation could not be inferred) and the groupings based on the phylogeny. (b) ML phylogenies of each major inversion estimated using IQ-TREE, based on mapping of resequence data to the H. melpomene reference. (c) Alignments of w2rap haplotype merged scaffolds to the H. melpomene reference genome supporting the inversion on chromosome 13. Inversion breakpoints are depicted by the vertical red lines. Scaffold alignments are represented by the arrows, the direction and color of the arrows representing whether the alignments are to the forward strand (yellow rightwards arrows) or the reverse strand (blue leftward arrows). Black arrows represent alignments spanning the inversion breakpoints. (d, e) fdM and relative node depth (RND) statistics along the genome. Both statistics were calculated in 25 kb nonoverlapping windows across the genome, based on mapping of resequencing data to the H. melpomene reference. Chromosomes are shown with alternating gray and black colors. The location of inversions is given by the dashed vertical lines whereas horizontal red lines represent ±3 SD from the mean fdM and RND values. Outlier windows overlapping the chromosome 13 inversion are indicated by the yellow arrows. Positive fdM values (and lowered RND) indicate an excess of shared variation between H. burneyi with H. erato and negative values of fdM represent an excess of shared with H. melpomene. In this test, H. melpomene and H. burneyi were considered to be the ingroup species and H. erato the inner outgroup. Derived alleles were determined using E. tales (et al.). Species codes are as in figure 1.