Abstract
Mitochondrial complex I is the main site for electron transfer to the respiratory chain and generates much of the proton gradient across the inner mitochondrial membrane. Complex I is composed of two arms, which form a conserved L-shape. We report the structures of the intact, 47-subunit mitochondrial complex I from Arabidopsis thaliana and the 51-subunit complex I from the green alga Polytomella sp., both at around 2.9 Å resolution. In both complexes, a heterotrimeric γ-carbonic anhydrase domain is attached to the membrane arm on the matrix side. Two states are resolved in A. thaliana complex I, with different angles between the two arms and different conformations of the ND1 (NADH dehydrogenase subunit 1) loop near the quinol binding site. The angle appears to depend on a bridge domain, which links the peripheral arm to the membrane arm and includes an unusual ferredoxin. We propose that the bridge domain participates in regulating the activity of plant complex I.
An unusual ferredoxin completes a protein bridge that links the two arms of plant mitochondrial complex I and adjusts their angle in an open or closed conformation.
Introduction
Complex I is the largest enzyme complex of the mitochondrial electron-transfer chain. The complex catalyzes electron transfer from NADH onto ubiquinone, and couples it to proton translocation across the inner mitochondrial membrane (Agip et al., 2019; Parey et al., 2020; Sazanov, 2015). A homologous enzyme complex is present in several proteobacteria. Bacterial and mitochondrial complex I consists of two modules: the membrane arm and the peripheral arm. The membrane arm is integral to the mitochondrial or bacterial inner membrane, while the peripheral arm protrudes into the mitochondrial matrix or the bacterial cytoplasm. Together, the two arms describe an l-shape. Each arm has two functional domains: the NADH oxidation (N) and ubiquinone reduction (Q) domains make up the peripheral arm; and the proximal (relative to the peripheral arm) and distal proton translocating (PP and PD) domains make up the membrane arm.
The first high-resolution structures of complex I were bacterial (Baradaran et al., 2013; Berrisford et al., 2016). E. coli complex I has a mass of ∼500 kDa and is composed of 14 protein subunits, 7 of which make up the membrane arm and another 7 the peripheral arm. This set of 14 conserved subunits forms the core of complex I. Electrons are transferred from NADH to ubiquinone via one flavin mononucleotide (FMN) and 7 FeS clusters in the peripheral arm. Two additional FeS clusters in the peripheral arm of E. coli complex I are thought to support electron transfer indirectly. The membrane arm has four potential proton translocation pathways. The molecular mechanisms that couple electron transfer to proton translocation are currently unknown but are thought to involve long-range conformational changes between and within the two complex I arms (Berrisford et al., 2016).
Mitochondrial complex I is significantly larger than E. coli complex I. Apart from the 14 core subunits, it contains ∼30 accessory subunits. The first higher-resolution structures of mitochondrial complex I were from the aerobic yeast Yarrowia lipolytica (Zickermann et al., 2015) and mammals (Fiedorczuk et al., 2016; Zhu et al., 2016). Recently, more detailed cryoEM (electron cryo-microscopy) structures were reported for fungal (Grba and Hirst, 2020; Parey et al., 2018) and mammalian complex I (Agip et al., 2018; Kampjut and Sazanov, 2020). Mammalian complex I has 45 subunits and a mass of ∼970 kDa. The accessory subunits surround the core subunits and are thought to stabilize the complex. Some accessory subunits are likely to add new functions to complex I. For instance, two copies of a mitochondrial acyl carrier protein (ACP; also called SDAP) are integral parts of the mammalian and yeast complex I. Furthermore, a nucleoside kinase is attached to complex I in mammals and a sulfur transferase to that of Yarrowia (D'imprima et al., 2016).
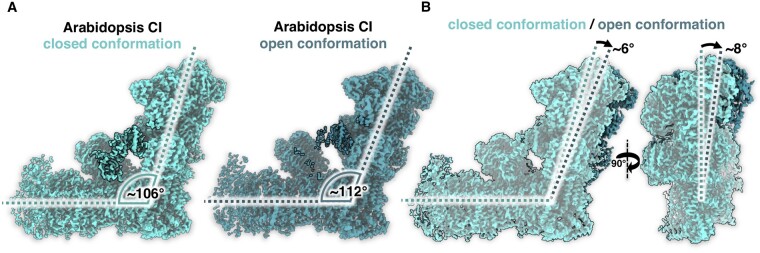
Structural investigation of mammalian complex I revealed several different conformations (Fiedorczuk et al., 2016; Zhu et al., 2016, reviewed in Agip et al., 2019). Two prominent conformations differ with respect to the angle between the peripheral and membrane arm, which is 112° in the open conformation and 105° in the closed conformation (Kampjut and Sazanov, 2020). Since the two conformations are associated with different loop structures near the Q reduction site, they were proposed to represent substates of the catalytic cycle. So far, open and closed conformations have not been reported for Yarrowia complex I (Parey et al., 2018, 2019; Grba and Hirst, 2020). The exact molecular mechanism by which complex I couples NADH oxidation to proton translocation remains to be elucidated.
Plants have two different versions of complex I, one each for chloroplasts and mitochondria. The chloroplast complex resembles that of cyanobacteria, which transfers electrons from ferredoxin to plastoquinone. The high-resolution structure of cyanobacterial complex I has recently been determined by cryoEM (Laughlin et al., 2019; Schuller et al., 2019; Zhang et al., 2020). The structure of plant mitochondrial complex I is less well characterized. Low-resolution single-particle EM revealed a second matrix-exposed domain near the center of the membrane arm (Dudkina et al., 2005). Plant complex I includes additional subunits not present in complex I from Yarrowia and mammals (Heazlewood et al., 2003), most notably proteins resembling γ-type carbonic anhydrases (γCAs; Parisi et al., 2004; Perales et al., 2004). The γCAs form a heterotrimer and were shown to account for the extra matrix-exposed domain (Sunderhaus et al., 2006; Fromm et al., 2016c). It has been proposed that the γCA domain is involved in the transfer of mitochondrial CO2 to the chloroplasts for carbon fixation (Braun and Zabaleta, 2007). First insights into the structure of this domain came from single-particle cryoEM of a complex I assembly intermediate from mung bean (Vigna radiata), which includes 30 of its 47 subunits (Maldonado et al., 2020).
Here we report the high-resolution cryoEM structures of complete complex I from the model plant Arabidopsis thaliana in the open and closed conformation and from the unicellular heterotrophic green alga Polytomella sp. in the closed conformation. We present structural and functional insights into plant-specific features of mitochondrial complex I, most notably a protein bridge, which links the peripheral arm to the membrane arm. The bridge appears to adjust the angle between the two complex I arms and may be involved in regulating complex I activity. A recent cryoEM study (Soufari et al., 2020) provides insights into the structure of complex I from cabbage, but the map is of lower resolution, shows only one conformation, and the complex is incomplete. In particular, it lacks the bridge domain.
Results and discussion
Structure of the intact Arabidopsis and Polytomella complex I
We used single-particle cryoEM to determine the structure of complete complex I of A. thaliana and Polytomella sp. Intact mitochondrial complex I from Arabidopsis was purified as described (Supplemental Figure S1, (Klodmann et al., 2010). Polytomella sp. complex I was isolated via a similar procedure (Supplemental Figure S2). For Arabidopsis complex I, a total of 459,177 CTF (contrast transfer function) corrected and polished particles from 12,768 micrographs recorded with a Titan Krios electron microscope equipped with a K3 direct electron detector resulted in a 3.41 Å cryoEM map (Supplemental Figures S3 and S4). Multibody refinement of the peripheral arm and the PP and PD domains of the membrane arm improved the resolution of the Arabidopsis complex to 2.91 Å. Further focused 3D classification and refinement separated the particle set into 42,096 2D-projections of a closed conformation at 3.48 Å resolution and 48,933 2D-projections of an open conformation at 3.46 Å resolution. For Polytomella complex I, a total of 61,708 CTF-refined and polished single particles picked from 1,652 micrographs, likewise collected with a K3 camera on a Titan Krios electron microscope, resulted in a 3.11 Å cryoEM map. Multibody refinement of the peripheral and membrane arms improved the resolution to 2.88 Å (Supplemental Figures S5 and S6). Complex I structures from the two organisms have the typical l-shape. The γCA domain, which is characteristic of plant mitochondrial complex I, shows up prominently on the matrix side in both maps. The membrane arm is connected by a conspicuous protein bridge near the γCA domain to the Q domain of the peripheral arm.
Subunit composition and complete model of Arabidopsis mitochondrial complex I
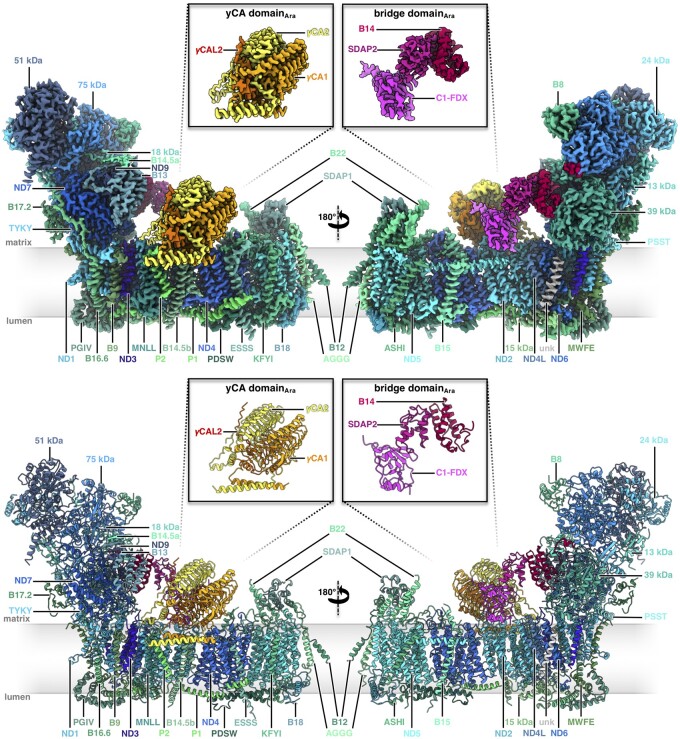
Arabidopsis complex I consists of 47 subunits (Figure 1, Supplemental Movie S1), including 14 core subunits and 33 accessory subunits (Table 1). For ease of comparison, we adopted the subunit nomenclature of bovine complex I (Walker et al., 1992), except for the 49 and 30 kDa subunits of the peripheral arm, which (unusually) are encoded by the mitochondrial genome in plants and are referred to as ND7 and ND9 (see Supplemental Table S1 for nomenclature). Of the 33 accessory subunits, 24 are conserved and found in both mammalian and plant complex I (Senkler et al., 2017a). Additionally, our structure indicates the presence of two copies of a mitochondrial ACP (mtACP1 and mtACP2, also known as SDAP1 and SDAP2) that were thought to be absent in plant complex I (Meyer et al., 2007), raising the number of conserved accessory subunits to 26. SDAP1 is located at the tip of the membrane arm; SDAP2 is part of the bridge domain (see below). The remaining seven accessory subunits are not present in fungal or mammalian complex I. These include (1) three members of the γCA/γCAL family (Fromm et al., 2016c); (2) the so-called P1 and P2 proteins (Meyer, 2012); (3) a small unknown hydrophobic subunit on the side of the membrane arm (Supplemental Figure S7); and (4) a ferredoxin, which we refer to as C1-FDX (complex I ferredoxin). The γCA/CAL, P1, and P2 proteins are known constituents of plant complex I. The small unknown hydrophobic subunit was also found in the cryoEM structure of the mung bean complex I intermediate (Maldonado et al., 2020). In contrast, the C1-FDX subunit has so far not been observed in plant complex I or any other complex I structure.
Figure 1.
Structure of mitochondrial complex I from Arabidopsis. Above: CryoEM density; below: atomic model. The 14 core subunits that are conserved in complex I in bacteria and eukaryotes are drawn in shades of blue; the accessory subunits in shades of green; the three subunits of the carbonic anhydrase domain in yellow, light orange and orange; subunits of the bridge domain in pink, purple, and red. Subunit nomenclature as for bovine complex I (Walker et al., 1992). Unknown (unk) refers to one subunit in the membrane arm that was not assigned due to limited amino acid sequence information. Figure insets: γ-carbonic anhydrase (γCA) and bridge domains. For EM statistics, see Supplemental Tables S2, S4, and S6.
Table 1.
|
Complex I subunits of Arabidopsis
|
Complex I subunits of Polytomella
|
||||
|---|---|---|---|---|---|
|
peripheral arm and bridge domain
|
peripheral arm and bridge domain
|
||||
| core subunits (7) |
conserved accessory subunits (9) |
plant-specific accessory subunits (1) |
core subunits (7) |
conserved accessory subunits (9) |
plant-specific accessory subunits (1) |
| 24 kDa | 13 kDa | C1-FDX | 24 kDa | 13 kDa | C1-FDX (NUOP3) |
| 51 kDa | 18 kDa | 51 kDa | 18 kDa | ||
| 75 kDa | 39 kDa | 75 kDa | 39 kDa | ||
| TYKY-1 | B8 | TYKY | B8 | ||
| PSST | B13 | PSST | B13 | ||
| ND7 | B14 | ND7 | B14 | ||
| ND9 | B14.5a | ND9 | B14.5a | ||
| B17.2 | B17.2 | ||||
| SDAP-2 | SDAP-2 | ||||
|
|
|
||||
|
membrane arm and γCA domain |
membrane arm and γCA domain |
||||
| core subunits (7) |
conserved accessory subunits (17) |
plant-specific accessory subunits (6) |
core subunits (7) |
conserved accessory subunits (18) |
plant-specific accessory subunits (9) |
|
|
|
||||
| ND1 | 15 kDa-1/2 | CA1/CA3 | ND1 | 15 kDa | CA1/CA3 |
| ND2 | AGGG | CA2 | ND2 | AGGG | CA2 |
| ND3 | ASHI | CAL2/CAL1 | ND3 | ASHI | CAL |
| ND4 | B9 | P1 | ND4 | B9 | NUOP4 |
| ND4L | B12-2 | P2 | ND4L | B12 | NUOP5 |
| ND5 | B14.5b | unknown su1 | ND5 | B14.5b | NUOP7 |
| ND6 | B14.7* | ND6 | B14.7 | NUOP8 | |
| B15 | B15 | unknown su1 | |||
| B16.6-2 | B16.6 | unknown su2 | |||
| B18 | B18 | ||||
| B22 | B22 | ||||
| ESSS-1 | ESSS | ||||
| KYFI | KYFI | ||||
| MNLL | MNLL | ||||
| MWFE | MWFE | ||||
| PDSW-1 | PDSW | ||||
| PGIV-2 | PGIV | ||||
| SDAP-1 | SDAP-1 | ||||
Subunit B14.7 (*) from Arabidopsis was not identified in the cryoEM density maps but has been detected by MS. Extensions -1/-2 indicate isoforms for some Arabidopsis complex I subunits. The dominant isoform was fitted to the map (except for SDAP, which occurs in two versions in Arabidopsis and Polytomella complex I). Unknown su: subunits not assigned due to limited amino acid sequence information. For protein accession numbers, see Supplemental Table S1.
Four subunits previously identified by mass spectrometry (MS) of Arabidopsis complex I (Supplemental Figure S8) were not found: (1) a l-galactono-1,4-lactone dehydrogenase; (2) a TIM–like protein (Arabidopsis accessions At1g18320 and At3g10110); (3) subunit B14.7; and (4) subunit SGDH. Of these, GLDH only binds to assembly intermediates of complex I (Schertl et al., 2012) and is not expected in the holocomplex, as recently confirmed by cryoEM (Soufari et al., 2020). The TIM-like protein may not be a true complex I subunit but was probably co-purified in earlier preparations. B14.7 is conserved in mammals, Yarrowia, and Polytomella, where its main role is thought to be in supercomplex formation (Letts et al., 2016; Kampjut and Sazanov, 2020). As a large fraction of Arabidopsis complex I forms a supercomplex with complex III2 (Eubel et al., 2003), the B14.7 subunit may have dissociated during complex I preparation. Neither the TIM-like protein nor subunit B14.7 were present in the reported cryoEM structures of complex I from mung bean and cabbage (Maldonado et al., 2020; Soufari et al., 2020). The SGDH subunit, like B14.7, is a conserved accessory subunit of mitochondrial complex I. Its location in mammalian complex I corresponds to that of the plant-specific P1 subunit in Arabidopsis. Mammalian SGDH and Arabidopsis P1 have a similar topology. We conclude that P1 is the plant equivalent of SGDH, as suggested (Soufari et al., 2020). In total, Arabidopsis complex I consists of 47 subunits plus subunit B14.7 (Table 1).
Apart from the 47 protein subunits, our structure of Arabidopsis complex I indicates the presence of 15 cofactors, including 11 in the peripheral arm (8 FeS clusters, 1 FMN, 1 Zn2+, and 1 NADPH) and four in the membrane arm (2 phosphopantetheine groups, 1 Zn2+, and 1 Fe ion), plus eight lipids and one LMNG detergent molecule (Supplemental Figure S9) in the membrane arm. NADPH is bound by the 39 kDa subunit of the peripheral arm that is not part of the NADH oxidation site. Binding of NADPH in this position is conserved in Yarrowia and mammals (Wirth et al., 2016); its function is currently unknown. The total molecular mass of Arabidopsis complex I including all subunits and cofactors is 1012 kDa.
Arabidopsis complex I was prepared without added substrates, and therefore the NADH oxidation site is empty, as expected. In contrast, the Q binding cavity contains a bound ubiquinone. Computational studies have postulated four possible Q binding positions (1, 1′, 2, 2′) in the binding cavity, of which sites 1 and 2 coincide with minima in the free energy profile of MD (molecular dynamics) simulations (Warnau et al., 2018). Both sites were confirmed by X-ray crystallography and CryoEM (for a review see Yoga et al., 2020a). Site 1 is within electron transfer distance of FeS cluster N2, while site 1’ is located 8–10 Å away from N2 towards the membrane surface. Sites 2 and 2’ are located ∼25 and 35 Å from N2 in the Q access pathway, close to the lipid bilayer. In the Arabidopsis complex I structure, a ubiquinone is bound at site 2’, ∼7 Å from site 2, where Q binds in the cryoEM structure of Yarrowia complex I (Parey et al., 2019).
Subunit composition and model of Polytomella mitochondrial complex I
The subunit composition of complex I from Polytomella sp. has been less well defined. We analyzed purified Polytomella complex I by 2D SDS/SDS PAGE and MS (Supplemental Figures S10–S13). In total, we identified peptides of 40 subunits with sequence similarity to known complex I components from other organisms, notably the closely related unicellular green alga Chlamydomonas reinhardtii. Shotgun MS analyses revealed another four polypeptides, raising the total number of detected subunits to 44 (Supplemental Figure S10; see also the GelMap at www.gelmap.de/2062). Since the MS data did not cover the complete amino acid sequences, especially those of the hydrophobic subunits, additional peptide sequence information was derived from genomes. The genome sequence of Chlamydomonas is known (Merchant et al., 2007). A partial genome sequence of Polytomella sp. has been determined recently (Murphy et al., 2019) but remains to be fully annotated. We used exon-intron prediction programs to identify open reading frames that encode the complete polypeptide sequences of complex I subunits in Polytomella.
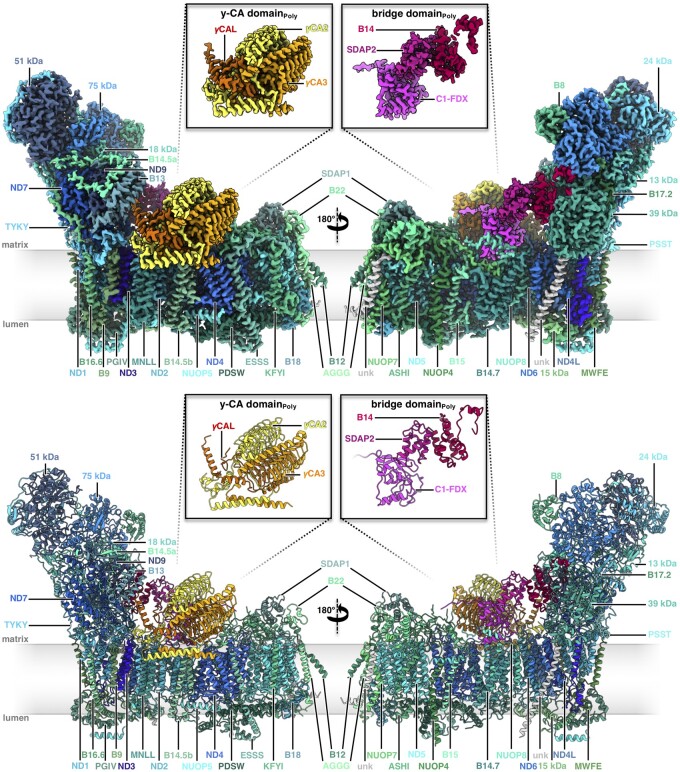
We were able to assign 49 proteins in the 2.9 Å cryoEM map of Polytomella complex I (Figure 2, Supplemental Movie S2). In addition, we discovered two proteins of unknown identity. All subunits detected by MS were found, including a second copy of the ACP (SDAP). Five subunits not detected by MS were identified directly on the basis of their cryoEM map density in combination with amino acid sequence information. Three of them are homologs of complex I subunits from other model species (Supplemental Figure S14). Two other proteins are new complex I subunits and were named NUOP7 and NUOP8 (Figure 2) in accordance with the nomenclature introduced for C. reinhardtii (Cardol et al., 2005). One of the two unassigned protein densities of Polytomella complex I corresponds to the small hydrophobic subunit at the side of the Arabidopsis membrane arm, the other one is an extra subunit located at the very tip of the membrane arm (Supplemental Figure S15). In summary, our Polytomella complex I density map includes 51 proteins: the complete set of the 14 core subunits and 37 accessory subunits (Table 1).
Figure 2.
Structure of mitochondrial complex I from Polytomella sp. Above: CryoEM density; below: atomic model. Color scheme for the 14 conserved core subunits, accessory subunits and the subunits of the carbonic anhydrase and bridge domains as in Figure 1. Insets: γ-carbonic anhydrase (γCA) and bridge domains. Unknown (unk) refers to two unassigned densities in the membrane arm with limited amino acid sequence information. For EM statistics, see Supplemental Tables S3, S5, and S7.
Of the 37 accessory subunits, 30 are conserved between Polytomella and Arabidopsis. Additionally, the small hydrophobic subunit at the side of the membrane arm is conserved, but its sequence is not known in either species. The plant-specific P1 and P2 subunits are absent in Polytomella. The six Polytomella subunits not present in Arabidopsis are B14.7, NUOP4, NUOP5, the new NUOP7 and NUOP8 proteins, and the unknown protein at the tip of the membrane arm. As in mammals and Yarrowia, the B14.7 subunit is found at the position where complex I interacts with the complex III dimer in the I+III2 supercomplex. NUOP4 and NUOP5 were detected by MS in Polytomella (Table 1, Supplemental Figures S10–S13) and previously in C. reinhardtii (Cardol et al., 2004, 2005).
The two acyl carrier subunits SDAP1 and SDAP2 in the Polytomella complex are in the same locations as in the Arabidopsis complex (Figure 2). Their sequences differ in a few residues. The Polytomella complex I ferredoxin C1-FDX (see below) is a homolog of the NUOP3 subunit previously identified in C. reinhardtii complex I (Cardol et al., 2004, 2005, 2008).
The overall structures of Arabidopsis and Polytomella complexes I are similar, including the trimeric γCA domain and the bridge connecting the Q domain of the peripheral arm to the membrane arm near the γCA domain. Polytomella complex I appears to be more compact, because it includes additional subunits. The Polytomella structure indicates 13 bound co-factors (8 FeS clusters, 1 FMN, 1 Zn2+, and 1 NADPH in the peripheral arm; two phosphopantetheine groups in the two acyl carrier subunits of the membrane arm), plus 12 lipid molecules (Supplemental Figure S16).
The γ-carbonic anhydrase heterotrimer of Arabidopsis and Polytomella complex I
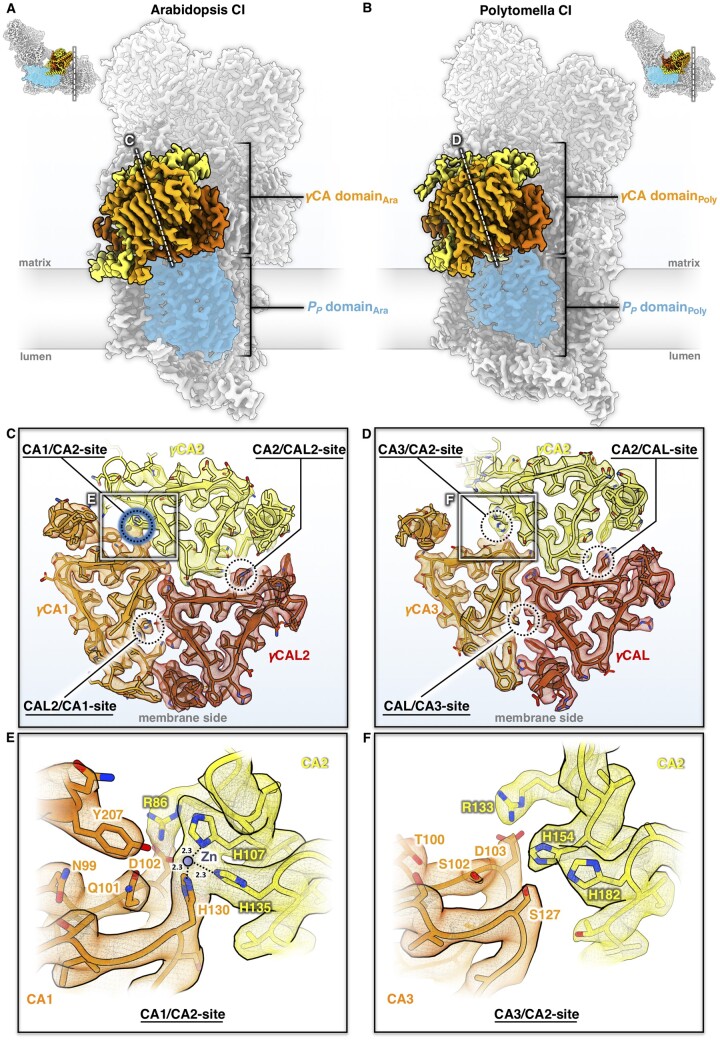
In plant mitochondria, a heterotrimeric γCA domain is attached to the membrane arm of complex I (Sunderhaus et al., 2006; Fromm et al., 2016c; Braun, 2020). The γCA domain is absent in complex I from mammals, fungi, bacteria, and chloroplasts but seems to occur in several groups of protists (Gawryluk and Gray, 2010). This suggests that the γCA proteins are of ancient origin and most likely formed part of complex I in the earliest ancestors of the eukaryotic clade. The Arabidopsis genome encodes five different γCA subunits, all of them associated with mitochondrial complex I (Klodmann et al., 2010). Of these, three have the conserved amino acid residues that form the active site, as in the prototypic γCA of the archaeon Methanosarcina thermophila (Kisker et al., 1996); these subunits are referred to as γCA1, γCA2, and γCA3 (Parisi et al., 2004). Two other subunits are known as γCA-like (γCAL) proteins: γCAL1 and γCAL2 (Perales et al., 2004). The complex I-integral γCA/γCAL proteins of Arabidopsis assemble into heterotrimers of two γCA proteins and one γCAL (Fromm et al., 2016c; Braun, 2020), but the precise composition of the trimers was unknown. Our complex I structure of Arabidopsis includes γCA2, γCA1, and γCAL2, as indicated by evaluating amino acid positions that differ between the γCA and γCAL proteins (Figure 3, A and C; Supplemental Figure S17). Note that the high-resolution map of the Arabidopsis complex suggests a mixed occupancy of the γCA/γCAL heterotrimer, because side chains of alternative but less abundant γCA or γCAL subunits can be fitted at some positions. This is in line with the finding that γCA and γCAL subunits can substitute for each other in Arabidopsis knockout lines (Fromm et al., 2016c).
Figure 3.
The γCA domains of Arabidopsis and Polytomella mitochondrial complex I. A and B, Position of the γCA domain on the membrane arm. Complex I (CI) is seen from the tip of the membrane arm and cut at the plane as indicated by a dashed white line in the small insets. Subunit color scheme as in Figures 1 and 2. The PP domain of the membrane arm is shaded in blue. C and D, Cross-sections of the γCA domains of Arabidopsis (C) and Polytomella (D) at the level of the catalytic sites at the γCA/CAL subunit interfaces, as indicated by dashed circles. In Arabidopsis, one of the three possible catalytic sites coordinates a metal ion (presumably zinc; blue) and is therefore potentially active, whereas none of the three sites of the Polytomella γCA domain are occupied, and therefore they cannot be active. E and F, Details of the catalytic site regions in Arabidopsis (E) and Polytomella (F) (white boxes in C and D). Amino acids are indicated by a one-letter code. For further details, see Supplemental Figures S17–S20.
However, not all permutations of γCA and γCAL subunits within the carbonic anhydrase heterotrimer seem to be possible. Apparently, γCA sites in the trimer cannot be occupied by γCAL proteins, and conversely, the γCAL site cannot be occupied by γCA. This conclusion becomes obvious when we evaluate the cryoEM structure of Arabidopsis complex I: The γCA subunits have a long amphiphilic N-terminal helix, which interacts laterally with the membrane arm, thus stabilizing the attachment of the γCA heterotrimer to complex I. This N-terminal helix is absent in γCAL1 and γCAL2. Our conclusion is supported by findings for Arabidopsis knockout mutants (reviewed in Senkler et al., 2017b): If the genes for γCA1 and γCA2 are deleted, complex I does not assemble, implying that γCAL1 or γCAL2 cannot substitute for γCA1 and γCA2 (Fromm et al., 2016b). Similarly, if the genes for γCAL1 and γCAL2 are deleted, plants are not viable, suggesting that γCA proteins cannot substitute for γCAL (Wang et al., 2012). Similar conclusions were drawn from the structures of mung bean and cabbage complex I (Maldonado et al., 2020; Soufari et al., 2020).
The fold of the γCA/CAL proteins is highly conserved, consisting of a central left-handed triangular β-helix, which is laterally flanked by a C-terminal α-helix (Supplemental Figure S18, A and B). The long amphiphilic α-helix at the N-terminus is a typical feature of γCA1 and γCA2, which is absent in γCAL1, γCAL2, and the prototypic γCA of M. thermophila. The two amphiphilic helices of γCA1 and γCA2 form a coiled coil parallel to the membrane arm on the matrix side of the inner mitochondrial membrane. A gap between the coiled coil and the membrane arm is occupied by a distinct set of lipids, which might contribute to attaching the γCA domain to complex I (Supplemental Figure S18C). The lipid composition in this region is not conserved in mammals and fungi (Parey et al., 2019; Bridges et al., 2020; Kampjut and Sazanov, 2020). The γCA domain interacts with the ND2 and B14.5b subunits, the plant-specific complex I protein P2 and C1-FDX (see below). Interaction of the γCA trimer and the membrane arm is restricted to the PP module. The γCA/γCAL subunits are part of early assembly intermediates; in their absence, assembly of plant mitochondrial complex I is arrested (Ligas et al., 2019).
The archaeal γCA of M. thermophila is a homotrimer with three active sites at the subunit interfaces, each with a zinc ion coordinated by three histidines, two of which belong to one subunit and the third to another neighboring subunit (Kisker et al., 1996). In Arabidopsis, only the γCA1/γCA2 interface has a complete set of active site histidines (γCA1_H130, γCA2_H107, and γCA2_H135). Together, these three side chains coordinate a metal (presumably Zn) ion in a non-peptide density (Figure 3, C and E). The nearby conserved sidechains γCA1_N99, γCA1_Q101, γCA1_D102, γCA1_Y207, and γCA2_R86 are crucial for stability and the catalytic mechanism (Iverson et al., 2000; Ferry, 2010), suggesting that the γCA1/γCA2 site in the Arabidopsis complex is active. So far, there is no direct evidence for carbonic anhydrase activity in complex I. However, binding of inorganic carbon to Arabidopsis γ-type carbonic anhydrase trimers has indirectly been shown by expressing these proteins in E. coli (Martin et al., 2009). The two other sites in the heterotrimer lack some of the conserved zinc-binding residues, do not show a non-peptide density, and are therefore presumably inactive. Similar findings were reported for mung bean and cabbage (Maldonado et al., 2020; Soufari et al., 2020).
In Polytomella, three γCA proteins and one γCAL protein were identified by MS and evaluation of the partial genome sequence. Based on sequence similarity to Arabidopsis, we refer to the Polytomella γCA subunits as γCA1, γCA2, and γCA3. Of these, γCA2 and γCA3 are present in Polytomella complex I, in addition to one copy of γCAL (Figure 3B and Supplemental Figure S19). As in Arabidopsis, the structure of Polytomella complex I indicates a degree of mixed occupancy of γCA subunits, because alternative, less abundant side chains can be fitted at some positions. It is therefore likely that a small fraction of Polytomella complex I contains γCA1, γCA2, and γCAL. The topological arrangement of the γCA/CAL subunits and the anchoring of the γCA domain by the coiled coil N-terminal amphipathic helices are very similar to those of Arabidopsis (Supplemental Figure S20). Surprisingly, none of the three potential catalytic sites at the subunit interfaces has the complete set of three zinc-coordinating residues (Figure 3, D and F). At the γCA2/γCA3 interface, the third histidine is substituted by γCA3_S127, and nearby residues that participate in catalysis have been replaced. Since none of the three potential catalytic sites show any density for a bound metal ion, we conclude that the Polytomella γCA domain is inactive.
In photosynthetically active organisms, carbonic anhydrases play a role in carbon assimilation, carbon concentration mechanisms, or pH stabilization. It has been suggested that the carbonic anhydrase of complex I is required for the transfer of carbon dioxide from mitochondria to the chloroplasts for carbon fixation in the Calvin-Benson cycle (Braun and Zabaleta, 2007). Since Polytomella, unlike its close relative C. reinhardtii, does not photosynthesize, it might not need this activity. The notion of an inactive mitochondrial complex I γCA domain in photosynthetically inactive species is supported by the finding that the three zinc-coordinating histidines of γCA are not conserved in the non-photosynthetic amoeboid protist Acanthamoeba castellanii (Gawryluk and Gray, 2010).
It has recently been reported that cyanobacterial complex I is involved in carbon transport and concentration (Schuller et al., 2020). However, the cyanobacterial carbonic anhydrase subunits belong to a different enzyme class, and they are attached to the PD domain at the tip of the membrane arm. Furthermore, one of the proton transfer pathways in the cyanobacterial PD domain appears to have adapted to CO2 transfer. In contrast, the γCA domain of Arabidopsis mitochondrial complex I is attached to the PP domain, and its active site is not close to a proton transfer path. Our structure thus does not suggest that plant mitochondrial complex I is directly involved in CO2 or bicarbonate transport across the inner mitochondrial membrane. Bicarbonate formed at the γCA domain might be exported from the mitochondrial matrix by transporters unrelated to complex I.
A ferredoxin bridge between the ubiquinone reduction and the γ-carbonic anhydrase domain
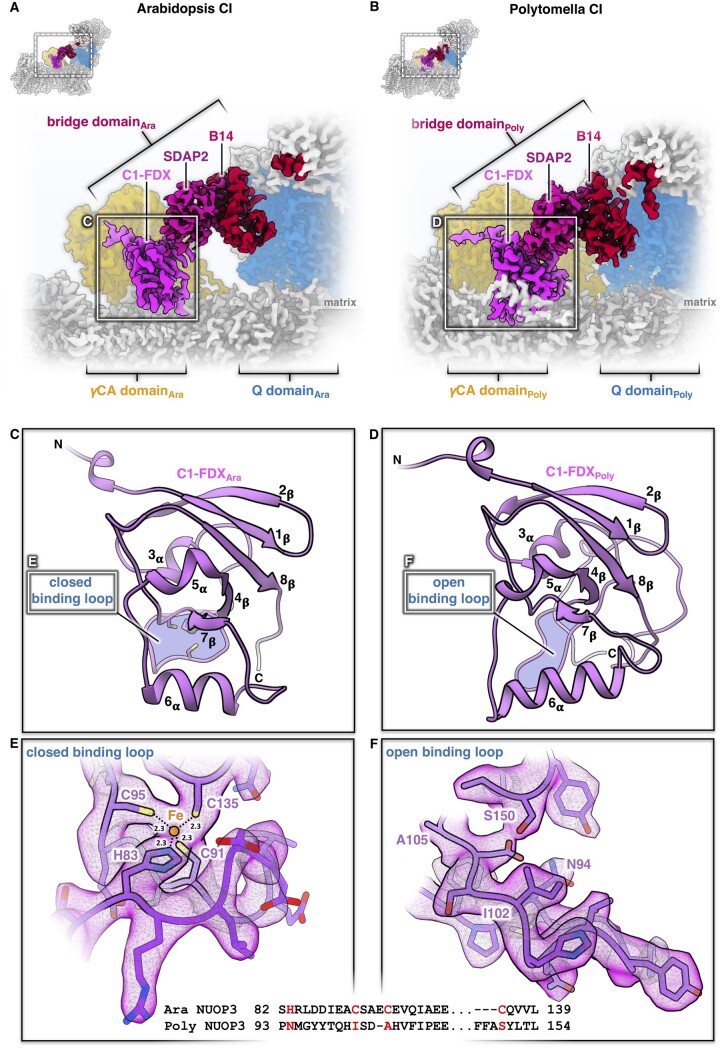
A striking feature of the Arabidopsis and Polytomella mitochondrial complex I is a three-subunit protein bridge, which forms a physical link between the Q domain of the peripheral arm and the membrane arm (Figure 4). The bridge consists of (1) subunit B14 in the peripheral arm, (2) one of the two ACPs (SDAP2, also called mtACP2) with an attached phosphopantetheine that binds a fatty acid of undefined chain length, and (3) a ferredoxin-like subunit (here referred to as complex 1 ferredoxin, C1-FDX). C1-FDX is connected to the core ND2 subunit of the membrane arm and the γCAL2 subunit of the γCA domain. The B14 and acyl carrier subunits are conserved in mammalian and Yarrowia complex I. The B14 subunit belongs to the eukaryotic LYR protein family (Angerer et al., 2014) that is defined by a (Leu, Tyr, Arg) motif close to its N-terminus. LYR proteins are comparatively small, mitochondria-specific, and positively charged components of respiratory chain complexes, or they act as assembly factors. ISD11, another LYR protein, is part of the iron-sulfur cluster (ISC) assembly complex. Mitochondrial ACPs are confined to the mitochondrial matrix (Angerer et al., 2017), where they are involved in fatty acid biosynthesis, in particular lipoic acid, which is a prosthetic group of several mitochondrial enzymes, and possibly also longer fatty acids for membrane biogenesis (Zensen et al., 1992). The two ACPs bound to complex I both carry longer fatty acids, which interact with the LYR-like subunits B14 and B22 (Fiedorczuk et al., 2016; Zhu et al., 2016). Finally, mitochondrial ACPs are known to form part of the ISC assembly complex, bind to ISD11, and might have a regulatory role in FeS cluster biosynthesis (Lill, 2020).
Figure 4.
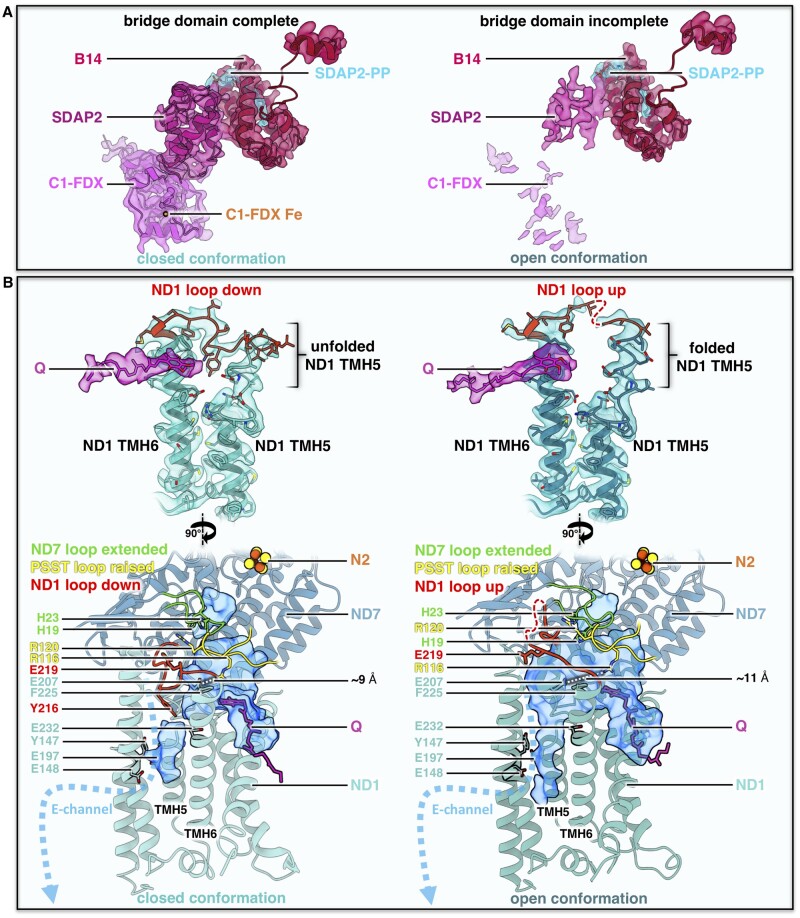
The bridge domain of Arabidopsis and Polytomella complex I (CI). A and B, Interaction of the bridge domain with the membrane arm near the γCA domain (orange) and the Q domain of the peripheral arm (blue). C and D, Structure of the C1-FDX subunit in Arabidopsis and Polytomella. E and F, catalytic sites of C1-FDX in Arabidopsis and Polytomella. For details, see Supplemental Figures S21 and S22.
Analysis of plant complex I by Blue-native/SDS PAGE did not reveal the C1-FDX subunit (e.g. Heazlewood et al., 2003; Meyer et al., 2008; Klodmann et al., 2011), but low levels of a ferredoxin were detected in complex I by complexome profiling, which combines one-dimension Blue-native PAGE with protein profiling by shotgun MS (Senkler et al., 2017b; Takabayashi et al., 2017). C1-FDX is clearly a part of our Arabidopsis complex I, which was prepared by sucrose density gradient ultracentrifugation. We conclude that Coomassie treatment of mitochondrial protein fractions during Blue-native PAGE destabilizes the bridge domain.
To further evaluate the interconnection of complex I and C1-FDX, we re-evaluated Arabidopsis mutants defective in genes encoding complex I-integral carbonic anhydrases. Proteome analyses of all investigated mutants revealed that the decrease in complex I levels correlates perfectly with C1-FDX levels: (1) Deletion of the γCA2 gene results in a roughly 80% loss of complex I and of the mitochondrial ferredoxin At3g07480 (Figure 8 in Perales et al., 2005), which we now identify as C1-FDX. (2) Similar results were obtained for a mutant that lacks γCAL1 and has lower amounts of γCAL2 (Table 1 in Fromm et al., 2016a). (3) Simultaneous deletion of γCA1 and γCA2 leads to a complete loss of complex I and a near-complete loss of C1-FDX (Supplemental Table 1 in Fromm et al., 2016b). Conversely, deletion of the gene encoding C1-FDX causes a reduction in complex I levels (Hansen et al., 2018). In wild-type Arabidopsis, the copy number of C1-FDX is estimated to be 2600 per single mitochondrion, as revealed by quantitative MS, which is in excellent agreement with the copy number of average mitochondrial complex I subunits (2500 per single mitochondrion; (Fuchs et al., 2020). Integrating available genetic, biochemical, and structural data, we conclude that C1-FDX is an accessory complex I subunit in Arabidopsis. Arabidopsis C1-FDX is homologous to NUOP3, which is part of Chlamydomonas complex I (Cardol et al., 2004, 2005). Polytomella NUOP3 (C1-FDX) is a structural subunit of the complex I bridge domain. We conclude that the bridge domain is a conserved structural feature of complex I in green algae and plants.
The 3D structure of Arabidopsis and Polytomella C1-FDX closely resembles mammalian and fungal mitochondrial ferredoxin 1 and 2, with its characteristic β-grasp fold (Supplemental Figure S21). Furthermore, C1-FDX resembles the mitochondrial ferredoxins of Arabidopsis known as AtMFDX1 and AtMFDX2 (Takubo et al., 2003). Although the degree of sequence identity is low, the structure of C1-FDX and the predicted structures of AtMFDX1 and AtMFDX2 modeled on human mitochondrial ferredoxin are very similar (Supplemental Figure S22). Mitochondrial ferredoxins are involved in the formation of FeS clusters (Lange et al., 2000; Cai et al., 2017). Typically, they have a central Fe2S2 cluster themselves, coordinated by four conserved cysteine residues in the core domain binding loop. In Arabidopsis C1-FDX, one of these cysteines is substituted by a histidine (Figure 4). Judging from the map density, the ligand bound by the four side chains of C1-FDX_H83, C1-FDX_C91, C1-FDX_C95, and C1-FDX_C135 cannot be a Fe2S2 cluster but must be a single metal ion, most likely iron.
Sites binding single iron ions are known from rubredoxins, which participate in electron transfer in various biological systems. In rubredoxins, the iron is coordinated by four conserved cysteins (Day et al., 1992; Bau et al., 1998). Another complex I subunit, the 13 kDa protein of the peripheral arm, may be related to proteins of the rubredoxin family (CL0045 in the Pfam database; Yip et al., 2011). The 13 kDa subunit binds Zn2+ by three cysteines and one histidine and is part of a superfamily of small zinc-finger proteins (Yip et al., 2011; Kmita et al., 2015). However, neither rubredoxins nor the 13 kDa subunit have a ferredoxin fold.
C1-FDX clearly does have a ferredoxin fold (Supplemental Figures S21 and S22), but its single metal ion site distinguishes it from all other known mitochondrial ferredoxins. The bound metal ion cofactor may be required for activity, although whether C1-FDX is active is currently unknown. In Polytomella, the C1-FDX equivalent NUOP3 lacks all four conserved metal-binding amino acids. As a result, the core domain loop is locked in a state that cannot bind a metal ion (Figure 4). We therefore assume that C1-FDX of Polytomella is inactive.
C1-FDX is localized at the side of the membrane arm of complex I, which binds to complex III2 in the I+III2 supercomplex. Apart from the B14.7 subunit of the membrane arm, C1-FDX may be involved in the physical contact of complex I with complex III2. Compared to I+III2 of fungi and mammals, this should increase the area of contact between the two complexes within the supercomplex. Indeed, the plant I+III2 supercomplex is particularly stable and therefore was the very first topologically defined respiratory supercomplex (Dudkina et al., 2005). It will be interesting to examine the structure of the I+III2 supercomplex from plant mitochondria.
Complex I ferredoxin sets the angle between the membrane and peripheral arm
Focused 3D classification and refinement of Arabidopsis complex I revealed two major sets of 2D projections that represent two conformations (Figure 5). The two structures differ in the angle between the two complex I arms, which is either 106° or 112°. Similar conformational classes have been described for mammalian complex I and are referred to as the open (angle 112°) and closed conformation (angle 105°; Kampjut and Sazanov, 2020; for review see Parey et al., 2020). At 3.5 Å resolution, the Arabidopsis 3D maps calculated for these conformations clearly differ with respect to the bridge domain (Figure 6A). In the open conformation, the acyl carrier and C1-FDX subunits are hardly visible and are either mostly dissociated or disordered. In contrast, both subunits are well defined in the closed conformation. Our data suggest that the acyl carrier and C1-FDX subunits of Arabidopsis complex I set the angle between its two arms. In Polytomella complex I, a helix of the NUOP8 subunit wraps around the lower part of the ferredoxin, connecting it firmly to the membrane arm (Figure 4B;Supplemental Figure S16), which would explain why the Polytomella complex is found exclusively in the closed conformation.
Figure 5.
Conformations of Arabidopsis complex I (CI). A, closed (angle 106°) and open (112°) conformation. B, Front and side views of superposed maps shown in A.
Figure 6.
Bridge domain and conformations of the Q binding site in the open and closed form of Arabidopsis complex I (CI). A, Map density and fitted atomic models of the bridge domain in Arabidopsis. Subunits color scheme as in Figures 1 and 2. B, Conformation of the Q binding site in the open and closed complex I conformations. Top: orientation of the loop (red) between TMH5 and TMH6 in ND1. Below: Q binding channel (blue) with E-channel on the left, showing the different conformations of ND1 together with the loop of ND7 (green, 49 kDa subunit in mammals) and PSST (yellow). Conserved amino acids drawn as sticks. Distances between ND1_E207 and PSST_R116 are indicated as dashed lines. For details, see Supplemental Figure S23.
The transition from the open to the closed conformation in Arabidopsis complex I is associated with a conformational switch of the loop linking transmembrane helices (TMH) 5 and 6 of the core ND1 (NADH dehydrogenase subunit 1) subunit in the membrane arm next to ubiquinone binding sites 2 and 2’. The ND1 loop is in a well-defined “down” orientation in the closed conformation of complex I, while the upper part of TMH 5 near the matrix surface of the membrane arm is unfolded (Figure 6B). In the open complex I conformation, the unfolded stretch folds into three turns of an α-helix, extending TMH 5 towards the matrix, and the loop switches from the “down” to an “up” configuration. The same switch of the ND1 loop has been reported for the ovine (sheep: Ovis aries) complex (Supplemental Figure S23A). In its catalytic cycle, ovine complex I has been suggested to alternate between the open and closed conformation (Kampjut and Sazanov, 2020). Both conformations are known to be present in active preparations of mammalian complex I (Letts et al., 2019). In contrast, the deactive state of mouse and bovine complex I was shown to adopt only the open conformation (Agip et al., 2018; Blaza et al., 2018). This open conformation was arrested by TMH 4 of the core ND6 subunit, which tilts by ∼35° to a new position on the external surface of the membrane arm. In Polytomella and in both conformations of the Arabidopsis complex I, this position is occupied by the TMH of the small unknown hydrophobic accessory subunit (Supplemental Figure S23E). Therefore, regulation of plant complex I must involve a different mechanism that locks the enzyme in the open conformation under substrate-depleted conditions.
In mammalian complex I, the transition from the open to the closed conformation is accompanied by distinct conformational changes of several subunit loops near Q reduction site 1. Changes in loop conformation are most striking at the distal end of the Q binding cavity and in the hydrophilic channel that branches off this cavity towards the so-called E-channel (Agip et al., 2018; Parey et al., 2019; Grba and Hirst, 2020; Kampjut and Sazanov, 2020). Apart from a major change in the ND1 loop (Figure 6B, Supplemental Figure S23A), detailed comparison of the closed and open conformations revealed only minor loop changes and helix twists of the Arabidopsis complex I (Supplemental Figure S23, B–D). In both conformations, the native quinol substrate resolved in our map binds in site 2’ near the Q channel entrance, with the Q headgroup mainly interacting with ND1_F225 (Figure 6B). Mutational studies and MD simulations have suggested that the disordered ND1 loop enables quinone diffusion within the cavity. Disordering thereby correlates with breaking salt bridges between the highly conserved glutamates of ND1 and arginines of the PSST loop (Yoga et al., 2019). In MD simulation of bovine complex I, breaking the salt bridge between ND1_E218 and PSST_R112 resulted in the diffusion of Q to site 2’ near the tunnel entrance, at a distance of ∼35 Å from FeS cluster N2.
In the open and closed conformation of the Arabidopsis complex I, distances of ∼11 Å and ∼9 Å between the corresponding sidechains of ND1_E207 and PSST_R116 do not allow a salt bridge to form (Figure 6B). The position of Q in site 2’ for both conformations is in perfect agreement with MD simulations (Yoga et al., 2019). The recent cryoEM structures of the ovine complex have shown that the formation of a salt bridge between ND1 and PSST is facilitated by a conformational change in the PSST loop itself (Kampjut and Sazanov, 2020). This change from the “flipped” to the “raised” position rotates the arginine of this potential salt bridge into close proximity to the ND1 glutamate. In Arabidopsis complex I, the PSST loop was found only in the raised position, but the observed distance between the arginine and glutamate sidechains would not allow a salt bridge to form (Figure 6B, Supplemental Figure S23B). Conformational changes in the ND1 loop are suggested to link the Q cavity with the E-channel by creating a water wire and enabling the transmission of an electrostatic pulse along the hydrophilic axis of the membrane arm for proton translocation (Zickermann et al., 2015; Grba and Hirst, 2020; Kampjut and Sazanov, 2020). Movement of the ND1 loop is also supposed to displace the ND3 TMH1-2 loop and in turn the ND7 β1-β2 loop (known as the 49 kDa loop in mammals; Cabrera-Orefice et al., 2018; Kampjut and Sazanov, 2020). Displacement of the ND7 β1-β2 loop would clear the path for the quinone to site 1 near the N2 FeS cluster, where it is reduced. However, inspection of the two conformations of the Arabidopsis complex I does not support the propagation of a structural change from ND1 to ND7. In both cases, the ND3 loop is disordered and the ND7 loop is extended, blocking access to Q binding site 1 (Figure 6B, Supplemental Figure S23, B and C). A detailed comparison of the two conformations of Arabidopsis complex I with the corresponding structures from mammals, fungi, and bacteria is complicated not only by the use of different substrates and inhibitors, but also different purification protocols.
In the future, structural studies in conjunction with functional assays should reveal whether there is a link between the observed differences in loop conformations at the Q binding site and the opening and closing of plant complex I. Mutational studies in Yarrowia have shown that interfering with the tight interaction between subunit NDUFA6 (B14 of the bridge domain) and the Q reduction domain caused the peripheral arm to tilt and structural elements in ND1, ND3, and ND7 to become disordered (Yoga et al., 2020b). Since C1-FDX is an integral part of the bridge joining the two arms, a looser binding or loss of the ferredoxin could also result in an opening of the l-shape and partial displacement of some loops at the matrix-exposed Q binding cavity.
Possible functions of the γCA and bridge domains
The structures of intact complex I from mitochondria of A. thaliana and Polytomella sp. show a heterotrimeric γCA domain attached to the membrane arm by coiled coil amphipathic helices of the two γCA subunits. Only one of the three catalytic sites of the γCA domain binds a metal ion in Arabidopsis, whereas none of them do in the chlorophyll-less alga Polytomella, implying that γCA in the Polytomella complex is inactive. This is in line with the hypothesis that the γCA domain promotes the transfer of mitochondrial CO2 for carbon fixation by the Calvin-Benson cycle to the chloroplasts (Braun and Zabaleta, 2007) and is therefore required only in photosynthetic organisms. The γCA domain is known to be essential for the assembly of plant mitochondrial complex I, but its connection to the newly discovered bridge domain in our structures suggests an additional role in controlling the mutual orientation of the two complex I arms, and possibly their activity.
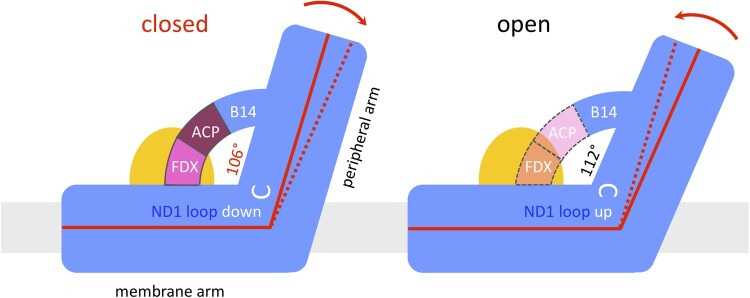
Of the three accessory subunits in the bridge domain, the peripheral arm protein B14 and the ACP (SDAP2) are conserved in mammals and Yarrowia. In the plant complex, the bridge is completed by the unusual ferredoxin C1-FDX. C1-FDX fills the gap between the ACP and the core subunit ND2 in the membrane arm close to the γCAL subunit of the carbonic anhydrase domain. The three-subunit protein bridge seems to set the angle between the two complex I arms (Figure 7). In the closed conformation of Arabidopsis and in Polytomella complex I, which is found only in the closed conformation, C1-FDX is firmly attached to the membrane arm. In Arabidopsis, disorder in the bridge domain or a loose attachment of C1-FDX to the membrane arm may enable the transformation from the closed into the open configuration. Although we cannot rule out the possibility that the bridge domain in some of the Arabidopsis particles had been damaged during isolation, the striking similarity of the two conformations of the Arabidopsis and ovine complex (which does not have the bridge) argues against this possibility.
Figure 7.
The ferredoxin bridge of Arabidopsis complex I. In the closed conformation, subunit B14 of the peripheral arm forms a bridge with the ACP (also designated SDAP2; purple) and the complex I ferredoxin C1-FDX (pink). Ferredoxin connects ACP with the membrane arm (ND2) near the gamma carbonic anhydrase domain (orange). The ND1 loop linking TMH5 and TMH6 of the ND1 subunit in the membrane arm (white) is down. The angle between the membrane and peripheral arms is set to 106°. In the open conformation, the bridge seems to be disrupted; the ACP (light pink) and F1-FDX (orange) subunits are disordered or (partially) absent; the ND1 loop flips to the “up” position, and the angle between the two complex I arms relaxes to 112°. Membrane, gray.
It is tempting to speculate that, as a redox-sensitive subunit, C1-FDX plays a role in the regulation of plant complex I activity. In the cryoEM structures of the ovine complex I, a positional switch of ND6 TMH 4 was thought to be responsible for locking the deactive form in the open, resting conformation (Kampjut and Sazanov, 2020). In complex I from Arabidopsis and Polytomella, this switch is prevented by the TMH of the small unknown hydrophobic subunit that occupies the position of the ovine ND6 TMH 4. This helix is clearly not part of ND6, which is well defined in the Polytomella map, and in Arabidopsis the ND6 sequence does not fit the helix density. A different regulatory mechanism must therefore apply to plant complex I. This mechanism may be related to C1-FDX in the bridge domain. The oxidation state of the metal ion bound by the ferredoxin might align complex I activity with the redox state of the mitochondrial matrix. Such a mechanism would be particularly appropriate for photosynthetic organisms.
Materials and methods
Plant material
A. thaliana was cultivated as described (Farhat et al., 2019). Briefly, Arabidopsis plants (ecotype Columbia 0) were grown under sterile conditions in a growth chamber (16 h light [sodium vapor lamps; 120 µmol photons m−2 s−1]/8 h dark, 22°C) for one week. Leaves were cut into small pieces and placed on B5 medium (3.16 g L−1 B5-medium, 3% sucrose [w/v], 0.75% agar [w/v], 0.5 mg L−1 2,4-d, 0.05 mg L−1 kinetin, pH 5.7) to induce callus formation. After three weeks, callus tissue was transferred into liquid B5 medium, which was refreshed once per week. The cell suspension culture was maintained at 22°C on a shaker in the dark.
Polytomella sp. cells (198.80, E.G. Pringsheim) were ordered from the SAG Culture Collection of Algae (Göttingen University, Germany) and cultivated in acetate medium (0.2% [w/v] sodium acetate, 0.1% [w/v] tryptone peptone, 0.1% [w/v] yeast extract) at 20°C in the dark; the medium was changed twice per week.
A. thaliana mitochondria isolation
Mitochondria were isolated from ∼150 g of A. thaliana suspension culture cells. The cells were harvested with a sieve. All subsequent steps were performed at 4°C or on ice. Cells were disrupted in grinding buffer (450 mM sucrose, 15 mM MOPS, 1.5 mM EGTA, 0.6% [w/v] PVP40, 0.2% [w/v] BSA, 10 mM sodium ascorbate, 10 mM cysteine, pH 7.4, 0.2 mM PMSF) using a Waring blender. The suspension was centrifuged at 2700 × g (twice) and 8300 × g for 5 min, respectively, to remove cell debris. Mitochondria were pelleted by centrifugation at 17,000 × g for 10 min, resuspended in washing buffer (300 mM sucrose, 10 mM MOPS, 1 mM EGTA, pH7.2, 0.2 mM PMSF), and carefully dispersed using a Dounce homogenizer. Isolated mitochondria were loaded onto discontinuous Percoll gradients (18%, 23%, and 40% Percoll in gradient buffer [300 mM sucrose, 10 mM MOPS, pH 7.2]). Percoll gradient ultracentrifugation was performed at 70,000 × g for 90 min. Mitochondria were collected from the 23%–40% interphase of the Percoll gradients. Percoll was removed by three cycles of pelleting the mitochondria by centrifugation at 14,500 × g for 10 min and resuspending the pellets in resuspension buffer (400 mM mannitol, 1 mM EGTA, 10 mM tricine, pH 7.2, 0.2 mM PMSF). Washed mitochondrial pellets were finally resuspended at a concentration of 0.1 g organelle pellet per milliliter resuspension buffer and stored at −80°C until use.
Purification of complex I from A. thaliana
Purified mitochondria from A. thaliana (containing ∼10 mg mitochondrial protein) were sedimented by centrifugation at 14,300 × g for 10 min at 4°C, resuspended in membrane solubilization buffer (30 mM HEPES, 150 mM potassium acetate, 1% [w/v] lauryl maltose neopentyl glycol [LMNG]), and incubated for 5 min on ice. Solubilized protein complexes were separated from membrane debris by centrifugation for 20 min at 18,300 × g and 4°C. Mitochondrial protein complexes were separated by sucrose gradient ultracentrifugation (Klodmann et al., 2010, modified). Sucrose gradients (volume: 15 mL) were prepared with a gradient mixer using 8 and 7 mL of a 1.5 M and 0.3 M sucrose solution (in 15 mM Tris, 20 mM KCl, 0.05% [w/v] LMNG, pH 7.0), respectively. One mg mitochondrial protein was loaded per gradient. Centrifugation was performed at 146,000 × g and 4°C for 20 h. The gradients were fractionated into 500 µL aliquots using a sample collector. To identify fractions containing complex I, 50 µL aliquots were analyzed by one-dimensional Blue-native PAGE (Wittig et al., 2006). Complex I was further purified by size-exclusion chromatography. Fractions containing complex I were pooled and loaded onto a Superose 6 Increase 10/300 column (GE Healthcare) equilibrated with buffer containing 30 mM HEPES-NaOH, pH 7.8, 50 mM KCl, and 0.007% [w/v] LMNG. Fractions containing complex I were concentrated using a Vivaspin 500 column with a 100,000 molecular weight cutoff. To remove sucrose, the concentrated sample was resuspended in size-exclusion buffer, concentrated to 1.1 mg mL−1, and used directly for cryoEM specimen preparation.
Purification of complex I from Polytomella sp.
Polytomella sp. mitochondrial complex I was purified following the protocol for the preparation of Polytomella ATP synthase (Murphy et al., 2019) with some modifications. Mitochondria (175 mg mitochondrial protein) harvested from a Polytomella culture in exponential growth phase were solubilized for 30 min at 4°C in a total volume of 12 mL buffer containing 30 mM Tris-HCl, pH 7.8, 50 mM NaCl, 2 mM MgCl2, and 2.9% [w/v] LMNG to a final detergent:protein weight ratio of 2:1. Unsolubilized material was removed by centrifugation at 21,000 × g for 15 min at 4°C. The supernatant was filtered and loaded onto a POROS GoPure HQ column (Thermo Fisher Scientific) connected to an Äkta purifier (GE Healthcare). The column was equilibrated in buffer A (30 mM Tris-HCl, pH 7.8, 50 mM NaCl, 2 mM MgCl2, 0.0085% [w/v] LMNG). After an initial wash with 100 mM NaCl in buffer A, complex I was eluted with a linear 100–300 mM NaCl gradient in buffer A. Fractions containing complex I were concentrated using an Amicon Ultra 4 column with 100,000 molecular weight cutoff and loaded onto a Superose 6 Increase 3.2/30 size-exclusion column (GE Healthcare). Complex I was eluted in buffer B (30 mM Tris-HCl, pH 7.4, 60 mM NaCl, 0.007% [w/v] LMNG) and used directly for cryoEM specimen preparation.
Analysis of the subunit composition of Polytomella complex I by two-dimensional SDS/SDS polyacrylamide gel electrophoresis (PAGE)
The 2D SDS/SDS PAGE of Polytomella complex I was carried out as described (Rais et al., 2004). Briefly, purified complex I from Polytomella sp. was mixed 1:1 with SDS sample buffer (10% [w/v] SDS, 30% glycerol [v/v], 100 mM Tris, 4% [v/v] mercaptoethanol, 0.006% [w/v] bromophenol blue, pH 6.8) and loaded onto a 10% polyacrylamide SDS gel containing 6 M urea. After first dimension SDS PAGE, a gel lane with separated subunits of complex I was excised, washed in acidic solution (100 mM Tris, 150 mM HCl, pH 2.0), and transferred horizontally onto a second dimension SDS gel (16% polyacrylamide, without urea). The 2D gels were stained with Coomassie blue (Neuhoff et al., 1985).
Protein analyses by MS
Protein spots were excised from 2D SDS/SDS gels. Proteins were fragmented into peptides by tryptic in-gel digestion as described (Klodmann et al., 2010). Tryptic peptide mixtures were analyzed by coupled liquid chromatography (LC)/electrospray (ESI)- quadrupole (Q)- time of flight (ToF) MS using the Easy nLC system (Thermo Scientific, Dreieich, Germany) and a micrOTOF Q II mass spectrometer (Bruker Daltonics, Bremen, Germany): Tryptic peptides were extracted (for details see Klodmann et al., 2010), resolved in solution P (0.1% formic acid, 2% acetonitrile in water), and transferred into the LC sample table. For peptide separation, a 2 cm C18 pre-column (ID 75µm, particle size 5µm, Thermo Scientific) and a 10 cm C18 analytical column (ID 75 µm, particle size 3 µm, Thermo Scientific) were used. A discontinuous elution gradient was applied by mixing solution A (0.1% formic acid in water) and solution B (0.1% formic acid in acetonitrile) as described (Klodmann et al., 2011). MS/MS parameters were applied as outlined before (Klodmann et al., 2011).
Evaluation of MS data
For protein identification, the following databases were searched with an in-house Mascot server: (1) a modified A. thaliana protein database, based on the TAIR database (www.arabidopsis.org) complemented with the edited sequences of mitochondrially encoded Arabidopsis proteins, (2) a Chlamydomonas reinhardtii database, and (3) a Polytomella database (both downloaded from NCBI in 10/2019). In addition, a Polytomella protein database translated from genomic DNA was used (Murphy et al., 2019). Finally, a database integrating all sequences of complex I subunits from all databases was built and used to evaluate MS data.
Shotgun MS
For shotgun MS, 50 µg purified Polytomella complex I was prepared by SDS PAGE and tryptic in-gel digestion as described (Thal et al., 2018). Extracted peptides were measured with an U3000 UPLC (Thermo Scientific, Dreieich, Germany) coupled to a Q Exactive Orbitrap MS system (Thermo Scientific, Dreieich, Germany) following a standard shotgun MS protocol (Thal et al., 2018).
Reference map for 2D-separated subunits of Polytomella complex I
A reference map of a 2D SDS/SDS gel of Polytomella complex I was established at the GelMap platform (www.gelmap.de) as described (Peters et al., 2013). The map (https://gelmap.de/2062) summarizes all MS-based identifications of complex I subunits from Polytomella.
Electron cryo-microscopy and image processing of A. thaliana complex I
A solution of 1.1 mg/mL purified complex I was applied onto C-flat 1.2/1.3 400 mesh copper grids (Science Services GmbH) that were glow-discharged for 45 s at 0.15 mA. The grids were frozen in liquid ethane after blotting for 4–7 s at blot force 20 using a Vitrobot operating at 10°C and 70% humidity.
Electron micrographs were collected at 300 kV in a Titan Krios G3i electron microscope equipped with a K3 detector operating in electron counting mode. The nominal magnification was 105,000×, giving a pixel size of 0.837 Å. 50-frame movies were recorded automatically with EPU software at an exposure rate of 15 electrons per pixel per second. Particles were picked using crYOLO, motion-corrected with MotionCor2, and the CTF was estimated with CTFFind4.1.13. Further processing was performed in Relion3. For initial 3D classification to clean the dataset, particles were binned to a pixel size of 2.511 Å. After 3D refinement with C1 symmetry applied to the whole complex, particles were re-extracted at a pixel size of 0.837 Å. Two additional rounds of CTF refinement and an intermediate step of Bayesian polishing resulted in a 3D reconstruction with an overall resolution of 3.41 Å. Further multibody refinement with a soft mask around the peripheral arm, the PP domain with the γCA and bridge domain, or the PD domain resulted in final resolutions of 2.99 Å, 3.06 Å, or 3.17 Å, respectively. To separate the closed and open conformations, particles were aligned to the peripheral arm with a local mask applied during 3D refinement, and then 3D-classified with a soft mask applied to the membrane arm with a value of T = 20 and without particle alignment. Particle classes were further refined with a global mask, resulting in a resolution of 3.53 Å for the closed conformation and 3.46 Å for the open conformation. Final focused 3D refinement around the PP, CA, bridge, and Q domain improved the resolution to 3.48 Å and 3.46 Å.
Electron cryo-microscopy and image processing of Polytomella sp. complex I
A solution of Polytomella complex I at a final concentration of 1.3 mg/mL was applied onto glow-discharged (0.15 mA for 45 s) C-flat 1.2/1.3 400 mesh copper grids (Science Services GmbH) and frozen in liquid ethane with a Vitrobot operating at 10°C and 70% with blot force 20 (6 s blotting time). Electron micrographs were collected at 300 kV with a Titan Krios G3i equipped with a K3 detector in electron counting mode. Fifty-frame movies were recorded automatically at a pixel size of 0.837 Å and an exposure rate of 15 electrons per pixel per second with EPU software. Movies were motion-corrected with MotionCor2, and the CTF was estimated with CTFFind4.1.13. Particles were picked using crYOLO. Processing was performed in Relion3. For the first two rounds of 3D classification to clean the dataset, particles were binned to a pixel size of 2.511 Å. After 3D refinement with C1 symmetry applied to the whole complex, particles were re-extracted at a pixel size of 0.837 Å. Two rounds of CTF refinement with an intermediate Bayesian polishing step and a subsequent 3D reconstruction resulted in an overall resolution of 3.11 Å. Additional multibody refinements with a soft mask around the peripheral and membrane arms resulted in final resolutions of 2.88 Å and 2.97 Å.
Model building
Initial models for the A. thaliana and Polytomella sp. complex I were built using homology models for each individual subunit created with the SWISS-MODEL server (Guex et al., 2009). Homology models were then rigid-body fitted into the cryoEM density maps using USCF Chimera (Pettersen et al., 2004), followed by manual building in Coot (Emsley et al., 2010). Final models were refined using the phenix.real_space_refine tool in Phenix (Afonine et al., 2018). Model quality statistics were taken from the phenix.validation_cryoem tool and are summarized in Supplemental Tables S2 and S3. For structural comparison, models were aligned using the Matchmaker tool of USCF Chimera. Water-accessible cavities were simulated with the program Hollow (Ho and Gruswitz, 2008) using an interior probe radius of 1.4 Å and a surface probe of 3.5 Å. Figures were drawn with UCSF Chimera and ChimeraX (Goddard et al., 2018).
Accession numbers
Proteomic data for Polytomella complex I are accessible via the GelMap platform at https://gelmap.de/2062. All structural data were submitted to the Protein Data Bank (PDB, https://www.rcsb.org/). For accession numbers, see Supplemental Table S4–S7.
Supplemental data
Supplemental Figure S1. Purification of complex I from Arabidopsis.
Supplemental Figure S2 . Purification of complex I from Polytomella.
Supplemental Figure S3. CryoEM of Arabidopsis complex I: Particle classification and processing scheme.
Supplemental Figure S4. CryoEM of Arabidopsis complex I: Orientation distribution and FSC curves.
Supplemental Figure S5. CryoEM of Polytomella complex I: Particle classification and processing scheme.
Supplemental Figure S6. CryoEM of Polytomella complex I: Orientation distribution and FSC curves.
Supplemental Figure S7. Unidentified helix density in Arabidopsis complex I.
Supplemental Figure S8. Subunit composition of Arabidopsis complex I, as revealed by 2D SDS/SDS PAGE.
Supplemental Figure S9. Cofactors and lipids in Arabidopsis complex I.
Supplemental Figure S10. Subunit composition of Polytomella complex I, as revealed by 2D SDS/SDS PAGE.
Supplemental Figure S11. Subunits of Polytomella complex I identified by mass spectrometry.
Supplemental Figure S12. Complex I of Polytomella: Peptides identified by coupled LC-ESI-MS/MS mass spectrometry.
Supplemental Figure S13. Partially assembled peptides of complex I subunits from Polytomella sp.
Supplemental Figure S14. Densities of Polytomella complex I assigned by homolog subunits.
Supplemental Figure S15. Non-assigned densities of Polytomella complex I.
Supplemental Figure S16. Cofactors and lipids of Polytomella complex I.
Supplemental Figure S17. γCA/γCAL proteins of the heterotrimeric γCA domain in Arabidopsis.
Supplemental Figure S18. Structures of γCA and γCAL subunits in Arabidopsis complex I.
Supplemental Figure S19. γCA/γCAL subunits in the heterotrimeric γCA domain of Polytomella.
Supplemental Figure S20. Structures of γCA and γCAL subunits in Polytomella complex I.
Supplemental Figure S21. Arabidopsis and Polytomella mitochondrial C1-FDX.
Supplemental Figure S22. Comparison of Arabidopsis C1-FDX to Arabidopsis mitochondrial FDX1 and FDX2.
Supplemental Figure S23. Peptide loops in the quinone reduction site of Arabidopsis complex I.
Supplemental Table S1. Nomenclature of complex I subunits in A. thaliana, C. reinhardtii, and other model species.
Supplemental Table S2. EM statistics for Arabidopsis thaliana complex I.
Supplemental Table S3. EM statistics for Polytomella sp. complex I.
Supplemental Table S4. Arabidopsis thaliana complex I map identifiers and statistics.
Supplemental Table S5. Polytomella complex I map identifiers and statistics.
Supplemental Table S6. Arabidopsis thaliana complex I model identifiers and quality statistics.
Supplemental Table S7. Polytomella complex I model identifiers and quality statistics.
Supplemental Movie S1. Three-dimensional cryoEM density map of complex I from A. thaliana at around 2.9 Å resolution.
Supplemental Movie S2. Three-dimensional cryoEM density map of complex I from Polytomella sp. at around 2.9 Å resolution.
Note: In our publication, the designations of mtACP-1 and mtACP-2 are flipped with respect to the nomenclature published by Meyer et al. 2007 (DOI 10.1007/s11103-007-9156-9).
Supplementary Material
Acknowledgments
We thank Janet Vonck and Volker Zickermann for critical comments on the manuscript.
Funding
This work was funded by the Max Planck Society (W.K., Ö.Y., and N.K.) and by the Deutsche Forschungsgemeinschaft (grant BR 1829/10-2—H.P.B. and J.S.; SFB 807—W.K. and N.K.).
Conflict of interest statement. None declared.
H.P.B. and W.K. initiated the project. N.K. purified complex I from Polytomella; N.K. and J.S. purified complex I from Arabidopsis. N.K. collected cryoEM data, performed image processing, and produced the figures. N.K. and Ö.Y. built and analyzed the atomic models. J.S. carried out proteome analysis. All authors evaluated data. H.-P.B. and W.K. wrote the manuscript, with contributions from N.K. and J.S.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Hans-Peter Braun (braun@genetik.uni-hannover.de) and Werner Kühlbrandt (werner.kuehlbrandt@biophys.mpg.de).
References
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD (2018) Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74: 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agip ANA, Blaza JN, Bridges HR, Viscomi C, Rawson S, Muench SP, Hirst J (2018) Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat Struct Mol Biol 25: 548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agip ANA, Blaza JN, Fedor JG, Hirst J (2019) Mammalian respiratory complex I through the lens of cryo-EM. Annu Rev Biophys 48: 165–184. [DOI] [PubMed] [Google Scholar]
- Angerer H, Radermacher M, Mańkowska M, Steger M, Zwicker K, Heide H, Wittig I, Brandt U, Zickermann V (2014) The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proc Natl Acad Sci U S A 111: 5207–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer H, Schonborn S, Gorka J, Bahr U, Karas M, Wittig I, Heidler J, Hoffmann J, Morgner N, Zickermann V (2017) Acyl modification and binding of mitochondrial ACP to multiprotein complexes. Biochim Biophys Acta Mol Cell Res 1864: 1913–1920. [DOI] [PubMed] [Google Scholar]
- Baradaran R, Berrisford JM, Minhas GS, Sazanov LA (2013) Crystal structure of the entire respiratory complex I. Nature 494: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bau R, Rees DC, Kurtz DM, Scott RA, Huang HS, Adams MWW, Eidsness MK (1998) Crystal structure of rubredoxin from Pyrococcus furiosus at 0.95 angstrom resolution, and the structures of N-terminal methionine and formylmethionine variants of Pf Rd. Contributions of N-terminal interactions to thermostability. J Biol Inorgan Chem 3: 484–493. [Google Scholar]
- Berrisford JM, Baradaran R, Sazanov LA (2016) Structure of bacterial respiratory complex I. Biochim Biophys Acta 1857: 892–901. [DOI] [PubMed] [Google Scholar]
- Blaza JN, Vinothkumar KR, Hirst J (2018) Structure of the deactive state of mammalian respiratory complex I. Structure 26: 312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun HP (2020) The oxidative phosphorylation system of the mitochondria in plants. Mitochondrion 53: 66–75. [DOI] [PubMed] [Google Scholar]
- Braun HP, Zabaleta E (2007) Carbonic anhydrase subunits of the mitochondrial NADH dehydrogenase complex (complex I) in plants. Physiol Plant 129: 114–122. [Google Scholar]
- Bridges HR, Fedor JG, Blaza JN, Di Luca A, Jussupow A, Jarman OD, Wright JJ, Agip ANA, Gamiz-Hernandez AP, Roessler MM. et al. (2020) Structure of inhibitor-bound mammalian complex I. Nat Commun 11: 5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Orefice A, Yoga EG, Wirth C, Siegmund K, Zwicker K, Guerrero-Castillo S, Zickermann V, Hunte C, Brandt U (2018) Locking loop movement in the ubiquinone pocket of complex I disengages the proton pumps. Nat Commun 9: 4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K, Tonelli M, Frederick RO, Markley JL (2017) Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron sulfur cluster biosynthesis. Biochemistry 56: 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P, Boutaffala L, Memmi S, Devreese B, Matagne RF, Remacle C (2008) In Chlamydomonas, the loss of ND5 subunit prevents the assembly of whole mitochondrial complex I and leads to the formation of a low abundant 700 kDa subcomplex. Biochim Biophys Acta 1777: 388–396. [DOI] [PubMed] [Google Scholar]
- Cardol P, Gonzalez-Halphen D, Reyes-Prieto A, Baurain D, Matagne RF, Remacle C (2005) The mitochondrial oxidative phosphorylation proteome of Chlamydomonas reinhardtii deduced from the genome sequencing project. Plant Physiol 137: 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P, Vanrobaeys F, Devreese B, Van Beeumen J, Matagne RF, Remacle C (2004) Higher plant-like subunit composition of mitochondrial complex I from Chlamydomonas reinhardtii: 31 conserved components among eukaryotes. Biochim Biophys Acta 1658: 212–224. [DOI] [PubMed] [Google Scholar]
- D'imprima E, Mills DJ, Parey K, Brandt U, Kühlbrandt W, Zickermann V, Vonck J (2016) Cryo-EM structure of respiratory complex I reveals a link to mitochondrial sulfur metabolism. Biochim Biophys Acta 1857: 1935–1942. [DOI] [PubMed] [Google Scholar]
- Day MW, Hsu BT, Joshuator L, Park JB, Zhou ZH, Adams MWW, Rees DC (1992) X-ray crystal-structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium pyrococcus-furiosus. Protein Sci 1: 1494–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP (2005) Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc Natl Acad Sci USA 102: 3225–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4): 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Jänsch L, Braun HP (2003) New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol 133: 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat N, Hichri S, Hildebrandt TM, Debez A, Braun HP (2019) Composition and stability of the oxidative phosphorylation system in the halophile plant cakile maritima. Front Plant Sci 10: 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry JG (2010) The gamma class of carbonic anhydrases. Biochim Biophys Acta 1804: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA (2016) Atomic structure of the entire mammalian mitochondrial complex I. Nature 538: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm S, Going J, Lorenz C, Peterhansel C, Braun HP (2016a) Depletion of the "gamma-type carbonic anhydrase-like" subunits of complex I affects central mitochondrial metabolism in Arabidopsis thaliana. Biochim Biophys Acta 1857: 1617–1618. [DOI] [PubMed] [Google Scholar]
- Fromm S, Senkler J, Eubel H, Peterhansel C, Braun HP (2016b) Life without complex I: proteome analyses of an Arabidopsis mutant lacking the mitochondrial NADH dehydrogenase complex. J Exp Bot 67: 3079–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm S, Senkler J, Zabaleta E, Peterhansel C, Braun HP (2016c) The carbonic anhydrase domain of plant mitochondrial complex I. Physiol Plant 157: 289–296. [DOI] [PubMed] [Google Scholar]
- Fuchs P, Rugen N, Carrie C, Elsasser M, Finkemeier I, Giese J, Hildebrandt TM, Kuhn K, Maurino VG, Ruberti C. et al. (2020) Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant J 101: 420–441. [DOI] [PubMed] [Google Scholar]
- Gawryluk RMR, Gray MW (2010) Evidence for an early evolutionary emergence of gamma-type carbonic anhydrases as components of mitochondrial respiratory complex I. BMC Evol Biol 10: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE (2018) UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grba DN, Hirst J (2020) Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation. Nat Struct Mol Biol 27: 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30: S162–S173. [DOI] [PubMed] [Google Scholar]
- Hansen BO, Meyer EH, Ferrari C, Vaid N, Movahedi S, Vandepoele K, Nikoloski Z, Mutwil M (2018) Ensemble gene function prediction database reveals genes important for complex I formation in Arabidopsis thaliana. New Phytologist 217: 1521–1534. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Howell KA, Millar AH (2003) Mitochondrial complex I from Arabidopsis and rice: orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochim Biophys Acta 1604: 159–169. [DOI] [PubMed] [Google Scholar]
- Ho BK, Gruswitz F (2008) HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson TM, Alber BE, Kisker C, Ferry JG, Rees DC (2000) A closer look at the active site of gamma-class carbonic anhydrases: High-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. Biochemistry 39: 9222–9231. [DOI] [PubMed] [Google Scholar]
- Kampjut D, Sazanov LA (2020) The coupling mechanism of mammalian respiratory complex I. Science 370: eabc4209. [DOI] [PubMed] [Google Scholar]
- Kisker C, Schindelin H, Alber BE, Ferry JG, Rees DC (1996) A left-handed beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. Embo J 15: 2323–2330. [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP (2011) Defining the protein complex proteome of plant mitochondria. Plant Physiol 157: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Sunderhaus S, Nimtz M, Jansch L, Braun HP (2010) Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita K, Wirth C, Warnau J, Guerrero-Castillo S, Hunte C, Hummer G, Kaila VR, Zwicker K, Brandt U, Zickermann V (2015) Accessory NUMM (NDUFS6) subunit harbors a Zn-binding site and is essential for biogenesis of mitochondrial complex I. Proc Natl Acad Sci USA 112: 5685–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Kaut A, Kispal G, Lill R (2000) A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc Natl Acad Sci USA 97: 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin TG, Bayne AN, Trempe JF, Savage DF, Davies KM (2019) Structure of the complex I-like molecule NDH of oxygenic photosynthesis. Nature 566: 411–414. [DOI] [PubMed] [Google Scholar]
- Letts JA, Fiedorczuk K, Degliesposti G, Skehel M, Sazanov LA (2019) Structures of respiratory supercomplex I+III2 reveal functional and conformational crosstalk. Mol Cell 75: 1131–1146. e1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts JA, Fiedorczuk K, Sazanov LA (2016) The architecture of respiratory supercomplexes. Nature 537: 644–648. [DOI] [PubMed] [Google Scholar]
- Ligas J, Pineau E, Bock R, Huynen MA, Meyer EH (2019) The assembly pathway of complex I in Arabidopsis thaliana. Plant J 97: 447–459. [DOI] [PubMed] [Google Scholar]
- Lill R (2020) From the discovery to molecular understanding of cellular iron-sulfur protein biogenesis. Biol Chem 401: 855–876. [DOI] [PubMed] [Google Scholar]
- Maldonado M, Padavannil A, Zhou L, Guo F, Letts JA (2020) Atomic structure of a mitochondrial complex I intermediate from vascular plants. Elife 9: e56664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Villarreal F, Miras I, Navaza A, Haouz A, Gonzalez-Lebrero RM, Kaufman SB, Zabaleta E (2009) Recombinant plant gamma carbonic anhydrase homotrimers bind inorganic carbon. FEBS Lett 583: 3425–3430. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al . (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH (2012) Proteomic investigations of complex I composition: how to define a subunit? Front Plant Sci 3: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Heazlewood JL, Millar AH (2007) Mitochondrial acyl carrier proteins in Arabidopsis thaliana are predominantly soluble matrix proteins and none can be confirmed as subunits of respiratory Complex I. Plant Mol Biol 64: 319–327. [DOI] [PubMed] [Google Scholar]
- Meyer EH, Taylor NL, Millar AH (2008) Resolving and identifying protein components of plant mitochondrial respiratory complexes using three dimensions of gel electrophoresis. J Proteome Res 7: 786–794. [DOI] [PubMed] [Google Scholar]
- Murphy BJ, Klusch N, Langer J, Mills DJ, Yildiz O, Kühlbrandt W (2019) Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F1-Fo coupling. Science 364: eaaw9128. [DOI] [PubMed] [Google Scholar]
- Neuhoff V, Stamm R, Eibl H (1985) Clear background and highly sensitive protein staining with coomassie blue dyes in polyacrylamide gels - a systematic analysis. Electrophoresis 6: 427–448. [Google Scholar]
- Parey K, Brandt U, Xie H, Mills DJ, Siegmund K, Vonck J, Kühlbrandt W, Zickermann V (2018) Cryo-EM structure of respiratory complex I at work. Elife 7: e39213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parey K, Haapanen O, Sharma V, Kofeler H, Zullig T, Prinz S, Siegmund K, Wittig I, Mills DJ, Vonck J, et al . (2019) High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease. Sci Adv 5: eaax9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parey K, Wirth C, Vonck J, Zickermann V (2020) Respiratory complex I - structure, mechanism and evolution. Curr Opin Struct Biol 63: 1–9. [DOI] [PubMed] [Google Scholar]
- Parisi G, Perales M, Fornasari M, Colaneri A, Schain N, Casati D, Zimmermann S, Brennicke A, Araya A, Ferry J, et al . (2004) Gamma carbonic anhydrases in plant mitochondria. Plant Mol Biol 55: 193–207. [DOI] [PubMed] [Google Scholar]
- Perales M, Eubel H, Heinemeyer J, Colaneri A, Zabaleta E, Braun HP (2005) Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I+III2 levels and alters mitochondrial physiology in Arabidopsis. J Mol Biol 350: 263–277. [DOI] [PubMed] [Google Scholar]
- Perales M, Parisi G, Fornasari M, Colaneri A, Villarreal F, Gonzalez-Schain N, Echave J, Gomez-Casati D, Braun HP, Araya A. et al. (2004) Gamma carbonic anhydrase like complex interact with plant mitochondrial complex I. Plant Mol Biol 56: 947–957. [DOI] [PubMed] [Google Scholar]
- Peters K, Belt K, Braun HP (2013) 3D Gel Map of Arabidopsis Complex I. Front Plant Sci 4: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rais I, Karas M, Schagger H (2004) Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification. Proteomics 4: 2567–2571. [DOI] [PubMed] [Google Scholar]
- Sazanov LA (2015) A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol 16: 375–388. [DOI] [PubMed] [Google Scholar]
- Schertl P, Sunderhaus S, Klodmann J, Grozeff GEG, Bartoli CG, Braun HP (2012) L-Galactono-1,4-lactone dehydrogenase (GLDH) forms part of three subcomplexes of mitochondrial complex I in Arabidopsis thaliana. J Biol Chem 287: 14412–14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller JM, Birrell JA, Tanaka H, Konuma T, Wulflhorst H, Cox N, Schuller SK, Thiemann J, Lubitz W, Setif P, et al . (2019) Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science 363: 257–260. [DOI] [PubMed] [Google Scholar]
- Schuller JM, Saura P, Thiemann J, Schuller SK, Gamiz-Hernandez AP, Kurisu G, Nowaczyk MM, Kaila VRI (2020) Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I. Nat Commun 11: 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Braun HP (2017a) Structure and function of complex I in animals and plants - a comparative view. Physiol Plant 161: 6–15. [DOI] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Eubel H, Hildebrandt T, Lengwenus C, Schertl P, Schwarzlander M, Wagner S, Wittig I, Braun HP (2017b) The mitochondrial complexome of Arabidopsis thaliana. Plant J 89: 1079–1092. [DOI] [PubMed] [Google Scholar]
- Soufari H, Parrot C, Kuhn L, Waltz F, Hashem Y (2020) Specific features and assembly of the plant mitochondrial complex I revealed by cryo-EM. Nat Commun 11: 5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderhaus S, Dudkina NV, Jänsch L, Klodmann J, Heinemeyer J, Perales M, Zabaleta E, Boekema EJ, Braun H-P (2006) Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J Biol Chem 281: 6482–6488. [DOI] [PubMed] [Google Scholar]
- Takabayashi A, Takabayashi S, Takahashi K, Watanabe M, Uchida H, Murakami A, Fujita T, Ikeuchi M, Tanaka A (2017) PCoM-DB update: a protein co-migration database for photosynthetic organisms. Plant Cell Physiol 58: e10. [DOI] [PubMed] [Google Scholar]
- Takubo K, Morikawa T, Nonaka Y, Mizutani M, Takenaka S, Takabe K, Takahashi MA, Ohta D (2003) Identification and molecular characterization of mitochondrial ferredoxins and ferredoxin reductase from Arabidopsis. Plant Mol Biol 52: 817–830. [DOI] [PubMed] [Google Scholar]
- Thal B, Braun HP, Eubel H (2018) Proteomic analysis dissects the impact of nodulation and biological nitrogen fixation on Vicia faba root nodule physiology. Plant Mol Biol 97: 233–251. [DOI] [PubMed] [Google Scholar]
- Walker JE, Arizmendi JM, Dupuis A, Fearnley IM, Finel M, Medd SM, Pilkington SJ, Runswick MJ, Skehel JM (1992) Sequences of 20 Subunits of NADH - ubiquinone oxidoreductase from bovine heart-mitochondria - application of a novel strategy for sequencing proteins using the polymerase chain-reaction. J Mol Biol 226: 1051–1072. [DOI] [PubMed] [Google Scholar]
- Wang Q, Fristedt R, Yu XH, Chen ZG, Liu HT, Lee Y, Guo HW, Merchant SS, Lin CT (2012) The gamma-carbonic anhydrase subcomplex of mitochondrial complex I is essential for development and important for photomorphogenesis of Arabidopsis. Plant Physiol 160: 1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnau J, Sharma V, Gamiz-Hernandez AP, Di Luca A, Haapanen O, Vattulainen I, Wikstrom M, Hummer G, Kaila VRI (2018) Redox-coupled quinone dynamics in the respiratory complex I. Proc Natl Acad Sci USA 115: E8413–E8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth C, Brandt U, Hunte C, Zickermann V (2016) Structure and function of mitochondrial complex I. Biochim Biophys Acta 1857: 902–914. [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H (2006) Blue native PAGE. Nat Protoc 1: 418–428. [DOI] [PubMed] [Google Scholar]
- Yip CY, Harbour ME, Jayawardena K, Fearnley IM, Sazanov LA (2011) Evolution of respiratory complex I: "supernumerary" subunits are present in the alpha-proteobacterial enzyme. J Biol Chem 286: 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoga EG, Angerer H, Parey K, Zickermann V (2020a) Respiratory complex I - Mechanistic insights and advances in structure determination. Biochim Biophys Acta 1861: 148153. [DOI] [PubMed] [Google Scholar]
- Yoga EG, Haapanen O, Wittig I, Siegmund K, Sharrna V, Zickermann V (2019) Mutations in a conserved loop in the PSST subunit of respiratory complex I affect ubiquinone binding and dynamics. Biochim Biophys Acta 1860: 573–581. [DOI] [PubMed] [Google Scholar]
- Yoga EG, Parey K, Djurabekova A, Haapanen O, Siegmund K, Zwicker K, Sharma V, Zickermann V, Angerer H (2020b) Essential role of accessory subunit LYRM6 in the mechanism of mitochondrial complex I. Nat Commun 11: 6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zensen R, Husmann H, Schneider R, Peine T, Weiss H (1992) Denovo synthesis and desaturation of fatty-acids at the mitochondrial acyl-carrier protein, a subunit of NADH-ubiquinone oxidoreductase in neurospora-crassa. Febs Lett 310: 179–181. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Shuai J, Ran ZX, Zhao JH, Wu ZF, Liao RJ, Wu J, Ma WM, Lei M (2020) Structural insights into NDH-1 mediated cyclic electron transfer. Nat Commun 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Vinothkumar KR, Hirst J (2016) Structure of mammalian respiratory complex I. Nature 536: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickermann V, Wirth C, Nasiri H, Siegmund K, Schwalbe H, Hunte C, Brandt U (2015) Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 347: 44–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.