Abstract
Although long non-coding (lnc)RNAs have been reported to be involved in the pathological development of bladder cancer, the functions of lncRNA prostate cancer-associated transcript 6 (PCAT6) and its underlying mechanism of action in bladder cancer remain unknown. The present study aimed to investigate the effect of PCAT6 in bladder cancer progression and explore its potential application as a novel treatment target. The expression of PCAT6 and miR-143-3p in bladder cancer tissues, adjacent normal tissues and cell lines was measured using reverse transcription-quantitative PCR. Fluorescence in situ hybridization assay was used to detect the subcellular localization of PCAT6. MTT, EdU, Transwell and wound healing assays were conducted to assess the biological function of PCAT6 on cell proliferation, migration and invasion. Putative binding sites between miR-143-3p and PCAT6 or PDIA6 were predicted using starBase, Lncbase and TargetScan analyzes. Dual-luciferase reporter assay was also used to confirm the potential binding between PCAT6 and miR-143-3p. RNA immunoprecipitation assay was performed to verify the possible interaction between PCAT6 and miR-143-3p. Western blotting was used to measure the expression of PDIA6. The results demonstrated that the expression levels of PCAT6 were upregulated in bladder cancer tissues relative to those in adjacent normal bladder tissues. Knockdown of PCAT6 served a role in suppressing the proliferation, migration and invasion of T24T and EJ bladder cancer cells. PCAT6 knockdown contributed to a reduction of PDIA6 expression at the mRNA and protein levels compared with that in negative control-transfected cells, whilst the miR-143-3p inhibitor partially mitigated this reduction effect. In addition, rescue experiments revealed that the miR-143-3p inhibitors reversed the effects of PCAT6 silencing on the malignant phenotypes of bladder cancer. Collectively, the results of the present study demonstrated that PCAT6 may serve an oncogenic role in bladder cancer via the miR-143-3p/PDIA6 axis. These results may provide a potential therapeutic target for the treatment of bladder cancer.
Keywords: long non-coding RNA, bladder cancer, prostate cancer-associated transcript 6, microRNA-143-3p, protein disulfide isomerase A6
Introduction
Bladder cancer is one of the most common urinary malignancies worldwide and has a high recurrence rate. In the United States, the numbers of new cases and deaths due to bladder cancer in 2019 were reported to be 80,470 and 17,670, respectively (1). Muscle-invasive bladder cancers represent 30% of all cases and are associated with a poor prognosis (2). Despite the recent progress in the treatment of bladder cancer, including surgery, radiation therapy, chemotherapy and immunotherapy, the prognosis of patients remains unsatisfactory, particularly for muscle-invasive and metastatic bladder cancer (3-5). The prognosis of bladder cancer is closely associated with the tumor stage; however, patients do not exhibit specific symptoms at the early stage (6). Therefore, identifying potential early detection markers is important for effective treatment.
Long non-coding (lnc)RNA represents a type of non-coding RNA >200 nucleotides long that lacks protein-coding capacity (7). To date, lncRNAs have been demonstrated to be associated with various intracellular and extracellular activities, such as gene transcription, mRNA splicing and tumorigenesis (8). In our previous studies, dysregulated lncRNAs, including growth arrest-specific 5 and sprouty4-intron 1, have been demonstrated to serve crucial roles in the development and progression of bladder cancer (9,10). However, the roles and the underlying mechanisms of the action of lncRNAs in the progression of bladder cancer remain unknown.
The prostate cancer-associated transcript 6 (PCAT6), a 1.0-kb intergenic lncRNA located at chromosome 1q32.1, was first detected in cervical cancer cells (11) and has been reported to be responsible for the regulation of radiosensitivity, cell proliferation and chemoresistance in various types of cancer, including breast (12), gastric (13) and cervical (14) cancer. However, the functions performed by PCAT6 in bladder cancer have not been reported to date. Therefore, the present study aimed to clarify the role of PCAT6 in the progression of bladder cancer and the potential underlying mechanism.
Materials and methods
Tissue specimens
A total of 32 pairs of bladder cancer and adjacent normal tissues (3 cm from the tumor margin) were obtained from patients who underwent radical cystectomy (age range, 41-84 years old; mean age, 62.1±11.7 years) at Wuhan Central Hospital (Wuhan, China) between January 2016 and September 2019. The patient clinical data are presented in Table I. Patients who had been treated with chemotherapy, laser therapy, cryotherapy or other anticancer modalities prior to the surgical procedure were excluded from the study. The specimens were classified according to the 2004 World Health Organization Consensus Classification and Staging System (15). The research protocol was approved by the Ethics Committee of the Central Hospital of Wuhan, and written informed consent was obtained from each patient before enrollment. The expression of PCAT6 in bladder cancer tissues was analyzed by Gene Expression Profiling Interactive Analysis (GEPIA 1.0; https://gepia.cancer-pku.cn/index.html) tools based on public data from the TCGA database (16).
Table I.
Associations between PCAT6 expression levels and the clinicopathological characteristics of patients with bladder cancer.
| Expression of PCAT6, n | ||||
|---|---|---|---|---|
| Parameter | Cases, n | Low | High | P-value |
| Sex | 0.3702 | |||
| Male | 23 | 19 | 4 | |
| Female | 9 | 6 | 3 | |
| Age | 0.6833 | |||
| <55 | 12 | 10 | 2 | |
| ≥55 | 20 | 15 | 5 | |
| Pathological stage | 0.0101 | |||
| pTa-T1 | 23 | 21 | 2 | |
| pT2-T4 | 9 | 4 | 5 | |
| Grade | 0.6317 | |||
| Low | 7 | 5 | 2 | |
| High | 25 | 20 | 5 | |
| Total | 32 | 25 | 7 | |
The specimens were classified according to the 2004 World Health Organization Consensus Classification and Staging System. PCAT6, prostate cancer-associated transcript 6.
Cell transfection
The human metastatic bladder cancer cell line T24T, which is a lineage-related lung metastatic variant of invasive bladder cancer cell line T24, was provided by Dr Dan Theodorescu (Departments of Urology, University of Virginia, Charlottesville, VA, USA) in 2010 and maintained at our laboratory as previously described (17-19). The cell line was subjected to DNA tests and authenticated in our previous study (20). Human bladder cancer cell lines EJ, UMUC3, 5637 and the human immortalized uroepithelium cell line (SV-HUC-1) were purchased from ATCC in 2015 and maintained at our laboratory. Cells were cultured in RPMI-1640 medium (HyClone; Cytiva) containing 10% fetal bovine serum (HyClone; Cytiva) in humidified air containing 5% CO2 at 37˚C. Small interfering (si)RNAs designed to target PCAT6 (siPCAT6), negative control siRNA (siNC), miR-143-3p mimic (miR-143-3p), miRNA mimic negative control (mimic-NC), miR-143-3p inhibitor (anti-miR143) and control inhibitor (control) were obtained from Guangzhou RiboBio Co., Ltd.
When the cell confluence reached 80%, a total of 100 nM siPCAT6, 100 nM siNC, 50 nM miR-143-3p mimics, 50 nM anti-miR143 or 50 nM of their corresponding negative controls were transfected into cells at 37˚C for 24 h using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) respectively. The sequences were as follows: siPCAT6, 5'-UGCAGCUCCGCUAUGGCCU-3'; siNC, 5'-UUCUCCGAACGUGUCACGUTT-3'; miR-143-3p, 5'-UGAGAUGAAGCACUGUAGCUC-3'; mimic-NC, 5'-UUCUCCGAACGUGUCACGUTT-3'; anti-miR143, 5'-GAGCUACAGUGCUUCAUCUCA-3'; inhibitor control, 5'-CAGUACUUUUGUGUAGUA-3'. After 48 h incubation at 37˚C, transfected cells were harvested and utilized for further experiments. The transfection efficiency was determined by RT-qPCR.
RNA extraction and reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse-transcribed to cDNA using the Prime Script™ RT Reagent kit (cat. no. RR037A; Takara Biotechnology Co., Ltd.). The temperature protocol using for RT was as follows: 37˚C for 15 min and 85˚C for 5 sec. For microRNA (miRNA) reverse transcription, miRNA First-Strand cDNA Synthesis kit (cat. no. B532451; Sangon Biotech Co., Ltd.) was used according to the manufacturer's instructions. qPCR was performed using SYBR® Premix Ex Taq (cat. no. RR041A; Takara Biotechnology Co., Ltd.) on the StepOnePlus Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Initial denaturation for 30 sec at 95˚C, followed by 40 cycles for 5 sec at 95˚C and 35 sec at 60˚C. GAPDH was used as an endogenous control for lncRNAs and mRNAs. The expression levels of miRNA were normalized to those of U6. The primers used were as follows: PCAT6 forward, 5'-CCCCTCCTTACTCTTGGACAAC-3' and reverse, 5'-GACCGAATGAGGATGGAGACAC-3'; miR-143-3p forward, 5'-GGGGTGAGATGAAGCACTG-3' and reverse, 5'-CAGTGCGTGTCGTGGAGT-3'; PDIA6 forward, 5'-GGAGGTCAGTATGGTGTTCAGGGAT-3'; and reverse, 5'-CTGCCACCTTGGTAATCTTCTGGTC-3'; GAPDH forward, 5'-TGCACCACCAACTGCTTAGC-3' and reverse, 5'-GGCATGGACTGTGGTCATGAG-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'.
Fluorescence in situ hybridization
Cy3-labeled mRNA probes for PCAT6 were designed and synthesized using cDNA as a template by Guangzhou RiboBio Co., Ltd (cat. no. lnc1cm001). Total RNA was isolated with RNAiso Plus (cat. no. 9109; Takara Biotechnology Co., Ltd.) from T24T cells and then reverse transcribed into cDNA with PrimeScript™ RT reagent Kit (cat. no. RR037A; Takara Biotechnology Co., Ltd.) as aforementioned. The probe sequence was is not publicly available. T24T and EJ cells were fixed in 4% paraformaldehyde for 30 min at 37˚C and incubated with 0.1% Triton X-100 on ice for 10 min. In total, 2.5 µl PCAT6 probes (20 µM) were hybridized with the cells for 5 h in the dark at 37˚C. Sodium citrate buffer was then added to wash the cell for 5 min at 42˚C for three times in the dark. 10 µl DAPI (5 µg/ml) was used to stain the nucleus for 10 min in the dark at room temperature. After washed with PBS for three times, images were captured using a Nikon A1Si Laser Scanning Confocal Microscope (magnification, x400; Nikon Instruments, Inc.).
MTT assay
The proliferative capacity of bladder cancer cells was evaluated by the MTT assay. At 48 h post-transfection, T24T and EJ cells were plated into 96-well plates at a density of 2,000 cells/well and incubated for 24, 48, 72 or 96 h. Subsequently, 20 µl MTT (5 mg/ml; EMD Millipore) was added in each well and cultured for 4 h. The culture medium was removed, and 150 µl DMSO was to dissolve the crystals. The absorbance was measured at 570 nm using a microplate reader.
5-Ethynyl-2'-deoxyuridine (EdU) assay
Transfected T24T and EJ cells (1x105 cells/well) were seeded in 96-well plates. EdU (100 mmol/l; Guangzhou RiboBio Co., Ltd.) was added to the medium and incubated for 2 h at room temperature in the dark. Subsequently, the DNA contents of the cells were stained with 100 µl Hoechst 33342 (2 µg/ml; Guangzhou RiboBio Co., Ltd.) for 30 min at room temperature in the dark. Images were captured with a fluorescent microscope (magnification, x200; Olympus Corporation). The ratio of EdU-stained cells to Hoechst-stained cells was calculated to evaluate the cell proliferation.
Wound healing assay
At 48 h post-transfection, T24T and EJ cells were seeded in a 6-well culture dish (5x105 cells/well) in RPMI-1640 medium (HyClone; Cytiva) containing 10% FBS (HyClone; Cytiva) at 37˚C. When the cell confluence reached 90%, cells were mechanically wounded by passing a sterile 200 µl plastic pipette tip through the monolayer with one stroke and incubated at 37˚C in serum-free medium. The cells were washed with PBS to wash away the floating cells. The wound healing processes were observed under a light microscope (magnification, x100; Olympus Corporation) at 0 and 48 h after the scratch, and the distance was analyzed with ImageJ software (National Institutes of Health). The relative migration rate (%)=(wound width at 0 h-wound width at 48 h)/wound width at 0 h x100.
Cell invasion and migration assay
Transwell chambers uncoated or coated with Matrigel (cat. no. 353097 for migration and cat. no. 354480 for invasion; 8-µm pore size filter; BD Biosciences) were used to determine the cell migratory or invasive abilities, respectively. Transfected T24T and EJ cells were seeded in the upper chamber of the Transwell insert with serum-free medium at a density of 1ⅹ104 cells/well. Medium supplemented with 10% fetal bovine serum was added to the lower chamber. Following 24-h incubation at 37˚C with 5% CO2, cells on the upper surface of the membrane were removed with a cotton swab. The migratory cells in the lower chamber were fixed with 4% paraformaldehyde for 15 min at room temperature and stained for 15 min at room temperature using 0.1% crystal violet. The numbers of migrated and invasive cells were counted under a light microscope (magnification, x100; Olympus Corporation) and the cell numbers were counted in five random fields of view.
Dual-luciferase reporter assay
The binding sites between miR-143-3p and PCAT6 were predicted and analyzed using Starbase 2.0 (https://starbase.sysu.edu.cn/) and Lncbase v.2 (https://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=site%2Findex). The binding sites between miR-143-3p and PDIA6 3'-untranslated regions (UTR) were predicted by TargetScan 7.1 (https://www.targetscan.org/vert_71/). The sequences of PCAT6 and PDIA6 3'UTR containing the potential wild-type (WT) or mutant (MUT) binding sites of miR-143-3p were synthesized by Tsingke Biological Technology Co., Ltd. and then inserted into the pGL3 vector (Guangzhou RiboBio Co., Ltd.) to construct the luciferase reporter. The T24T and EJ cells were seeded at a density of 1x104 into 96-well plates. The luciferase reporter (0.1 µg) and miR-143-3p mimics (40 nM) or mimic-NC (40 nM) were co-transfected into cells using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h of incubation at 37˚C, the transfected cells were harvested and assayed for luciferase activity using the dual-luciferase reporter assay system (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP) assay
The EZMagna RIP kit (cat. no. 17-701; EMD Millipore) was used to evaluate the target relationship between miR143-3p and PCAT6. T24T and EJ cells were harvested, resuspended with RIP lysis buffer supplemented with RNase Inhibitor (Promega Corporation) on ice for 5 min and centrifuged at 22,000 x g at 4˚C for 10 min. In total, 40 µl protein A/G beads and 5 µg human anti-Ago2 antibody (cat. no. ab32381; Abcam) or 5 µg negative control normal IgG (cat. no. ab188776; Abcam) were incubated at 4˚C for 8 h in 900 µl RIP buffer, before 2 mg total protein in 100 µl supernatant was added and incubated at 4˚C overnight. After brief centrifugation at 1,000 x g for 2 min at 4˚C, the samples were placed on a magnetic rack for 30 min at 4˚C. The supernatant was discarded and 500 µl RIP wash buffer was added to resuspend the beads, which was repeated five times. The supernatant was removed and samples were treated with proteinase K (cat. no. ST532; Beyotime Institute of Biotechnology.) on a shaker at 58˚C for 30 min. After centrifugation at 1,000 x g for 5 min at 4˚C, the supernatant was collected and 250 µl RIP buffer was added. A total of 400 µl mixture (phenol: Chloroform: Isoamyl alcohol, 125:24:1) was added to isolate the immunoprecipitated RNAs and the purified RNAs were subjected to RT-qPCR analysis.
Western blot assay
T24T and EJ cells were collected at 48 h post-transfection and resuspended in RIPA lysis buffer (Beyotime Institute of Biotechnology). The concentration of protein was determined using a bicinchoninic acid Protein Assay kit (Beyotime Institute of Biotechnology). In total, 50 µg of each protein samples were separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (EMD Millipore). Following blocking with 5% skimmed milk (Beyotime Institute of Biotechnology) for 2 h at room temperature, the membranes were incubated with primary antibodies against PDIA6 (1:2,000; cat. no. ab227545; Abcam) or GAPDH (1:5,000; cat. no. ab9485; Abcam) at 4˚C overnight. After being washed three times in TBS-T (0.1% Tween-20), the membrane was incubated with the HRP-conjugated Goat Anti-Rabbit IgG H&L secondary antibody (1:10,000; cat. no. ab97051; Abcam) at room temperature for 1 h before developing with an ECL kit (Beyotime Institute of Biotechnology). Data analysis was performed using ImageJ Software version 1.8.0 (National Institutes of Health) to evaluate the expression levels of proteins.
Statistical analysis
Data are presented as the mean ± standard deviation. Statistical analysis was performed by SPSS 17.0 (SPSS, Inc.) and GraphPad Prism 7.0 (GraphPad Software, Inc.). All experiments were independently repeated in triplicate. χ2 test was applied to determine the associations between PCAT6 expression and the clinical parameters of patients with bladder cancer. Pearson correlation analysis was used to analyze correlation between the expression. Comparison between tumor and adjacent non-cancerous tissues was analyzed using a paired Student's t-test. Comparison between two independent groups was analyzed via unpaired Student's t-test, whilst comparisons among multiple groups were calculated by one-way ANOVA followed by Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
PCAT6 is upregulated in bladder cancer tissues and cell lines
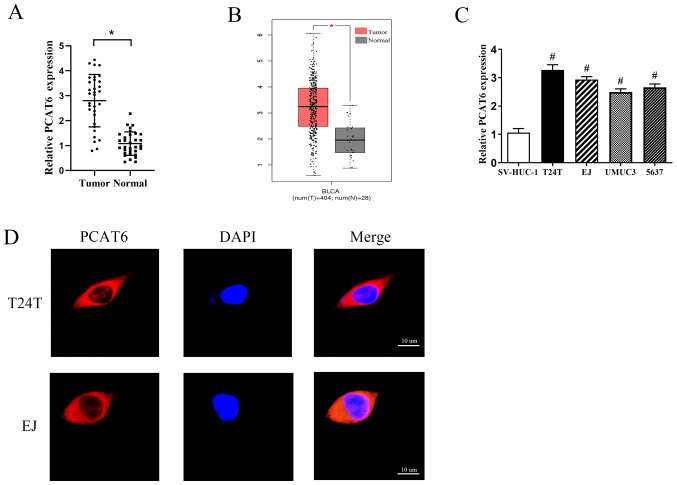
In order to determine the functions of PCAT6 in bladder cancer, the expression levels of PCAT6 in 32 pairs of bladder cancer and adjacent normal tissues were first determined. The results demonstrated that the expression levels of PCAT6 were markedly increased in bladder cancer tissues compared with those in the matched adjacent normal tissues (Fig. 1A). This was consistent with data from GEPIA (Fig. 1B). Additionally, the expression levels of PCAT6 were associated with the pathological stage, but not with other parameters such as sex, age or tumor grade (Table I). Subsequently, the expression levels of PCAT6 in four bladder cancer cell lines (T24T, EJ, UMUC3 and 5637) and one human normal bladder epithelial cell line (SV-HUC-1) were detected; as demonstrated in Fig. 1C, the levels of PCAT6 expression were significantly higher in the bladder cancer cells compared with those in SV-HUC-1 cells. Two cell lines, T24T and EJ, that exhibited the highest levels of PCAT6 expression were selected for further experiments.
Figure 1.
PCAT6 is upregulated in bladder cancer tissues and cell lines. (A) RT-qPCR was performed to determine the levels of PCAT6 expression in 32 pairs of bladder cancer and adjacent normal tissues. (B) PCAT6 levels in bladder cancer and normal tissues were analyzed using GEPIA database. (C) The expression levels of PCAT6 in four bladder cancer cell lines (T24T, EJ, UMUC3 and 5637) and a normal bladder epithelial cell line (SV-HUC-1) were determined by RT-qPCR. (D) The subcellular localization of PCAT6 was analyzed by fluorescent in situ hybridization. Scale bar, 10 µm. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 vs. tumor; #P<0.05 vs. SV-HUC-1. PCAT6, prostate cancer-associated transcript 6; RT-qPCR, reverse transcription-quantitative PCR.
Emerging studies have suggested that lncRNAs function through various regulatory mechanisms depending on their specific location in the cell (21,22). Therefore, the subcellular localization of PCAT6 in T24T and EJ cells was analyzed by fluorescent in situ hybridization. The results revealed that PCAT6 was mainly located in the cytoplasm (Fig. 1D).
Knockdown of PCAT6 suppresses bladder cancer cell proliferation, migration and invasion
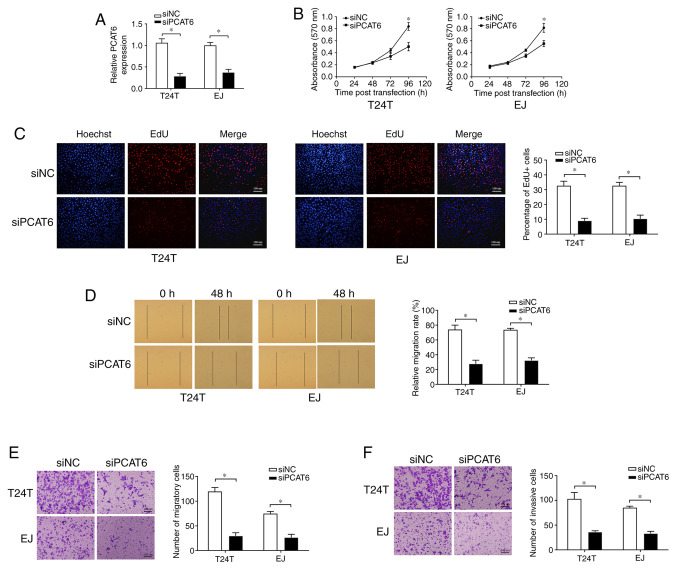
To determine whether PCAT6 may affect the malignancy of bladder cancer, T24T and EJ cells were transfected with siPCAT6, and RT-qPCR analysis demonstrated that the expression of PCAT6 was effectively inhibited (Fig. 2A). MTT and EdU assays were conducted to detect cell proliferation, which revealed that the knockdown of PCAT6 was suppressed cell proliferation compared with that in the siNC group (Fig. 2B and C). Subsequently, wound healing and Transwell assays were performed to determine the effects of PCAT6 on bladder cancer cell migratory capacity. As presented in Fig. 2D and E, knockdown of PCAT6 contributed to a sharp decline in the migration of T24T and EJ cells compared with that observed in the corresponding siNC groups. In addition, PCAT6 depletion also contributed to the significant suppression of cell invasion compared with that in the siNC groups, as demonstrated by Transwell invasion assay (Fig. 2F). Taken together, these results demonstrated that PCAT6 may serve an oncogenic role in bladder cancer.
Figure 2.
Knockdown of PCAT6 suppresses bladder cancer cell proliferation, migration and invasion. (A) The expression levels of PCAT6 following transfection of siPCAT6 or siNC were determined by reverse transcription-quantitative PCR. (B) MTT and (C) EdU assays were performed to detect cell proliferation after the knockdown of PCAT6. Cell migration ability was examined by (D) wound healing and (E) Transwell assays. Scale bar, 100 µm. (F) Cell invasive ability was analyzed by Transwell assay with Matrigel. Scale bar, 100 µm. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 vs. siNC. si, small interfering RNA; NC, negative control; PCAT6, prostate cancer-associated transcript 6.
PCAT6 acts as a sponge for miR-143-3p in bladder cancer cells
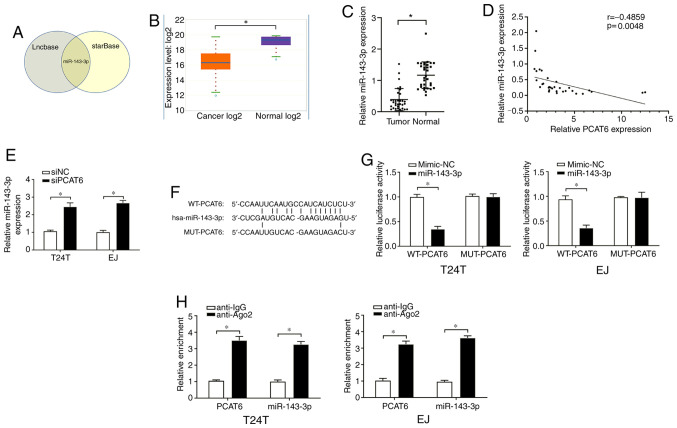
To predict the possible downstream targets of PCAT6, the online bioinformatics tools starBase and Lncbase were used and the results demonstrated that miR-143-3p contained a binding site for PCAT6 (Fig. 3A). starBase was further used to determine that the expression levels of miR-143-3p were significantly downregulated in bladder cancer tissues compared with those in the adjacent normal tissues (Fig. 3B). These results were confirmed by RT-qPCR assay in tissues from the patients included in the present study (Fig. 3C). In addition, Pearson's correlation analysis revealed a fair negative correlation between the expression levels of PCAT6 and miR-143-3p in the bladder cancer tissues (Fig. 3D). In addition, higher levels of miR-143-3p were observed in T24T and EJ cells following PCAT6 knockout compared with those in the siNC-transfected cells (Fig. 3E).
Figure 3.
PCAT6 acts as a sponge of miR-143-3p. (A) miR-143-3p was the only common target of PCAT6 predicted by Lncbase and starBase online tools. (B) The expression of miR-143-3p in bladder cancer and normal tissues in starBase. (C) miR-143-3p levels in bladder cancer and adjacent normal tissues were detected by qRT-PCR. (D) The correlation between PCAT6 and miR-143-3p was determined by Pearson's correlation analysis. (E) The expression levels of miR-143-3p were determined after the inhibition of PCAT6. (F) Putative binding sites between miR-143-3p and PCAT6 was identified using starBase. (G) Relative luciferase activities of bladder cancer cells following co-transfection with WT or MUT PCAT6 reporter plasmid and miR-143-3p mimics or NC mimics were detected. (H) Interaction between miR-143-3p and PCAT6 was further confirmed by RIP assay. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05. miR, microRNA; si, small interfering RNA; NC, negative control; PCAT6, prostate cancer-associated transcript 6; WT, wild-type; MUT, mutant.
To confirm the relationship between PCAT6 and miR-143-3p, a dual-luciferase reporter assay was performed. The WT and MUT sequences of PCAT6 were designed, and their binding sites to miR-143-3p are presented in Fig. 3F. As demonstrated in Fig. 3G, transfection with the miR-143-3p mimics induced a significant inhibition of the relative luciferase activity of WT-PCAT6, but not MUT-PCAT6, compared with that in the corresponding mimic-NC groups. Furthermore, the RIP assay revealed that PCAT6 and miR-143-3p were enriched in the Ago2-containing immunoprecipitants from T24T and EJ cells compared with the control IgG groups (Fig. 3H). These results suggested that miR-143-3p was a direct target of PCAT6 and negatively modulated by PCAT6.
PCAT6 upregulates the expression of PDIA6 via miR-143-3p
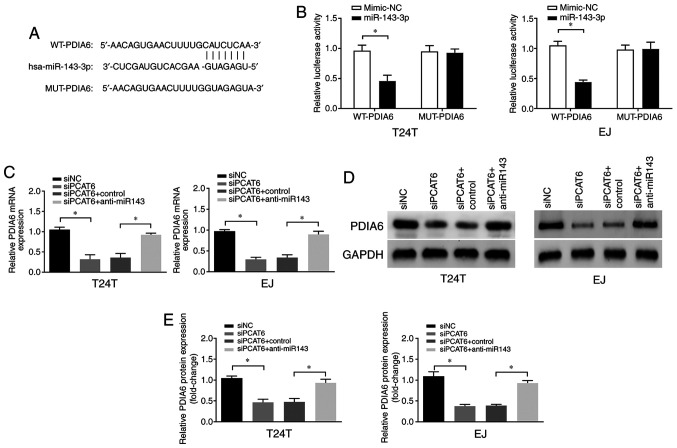
In order to determine the underlying mechanism of miR-143-3p-mediated progression of bladder cancer, the downstream targets of miR-143-3p were predicted using TargetScan software. As presented in Fig. 4A, PDIA6 contained a putative target sequence for miR-143-3p in its 3'-untranslated region. Therefore, we hypothesized that PCAT6 may exert its roles in bladder cancer via the miR-143-3p/PDIA6 axis. To verify this hypothesis, dual-luciferase reporter assay was performed, and the results demonstrated that the luciferase activity was suppressed following co-transfection of WT-PDIA6 and miR-143-3p compared with that in cells transfected with the mimic-NC (Fig. 4B), indicating that PDIA6 was a direct target of miR-143-3p in bladder cancer cells. In addition, PCAT6 knockdown led to a significant reduction of PDIA6 mRNA and protein expression levels, whereas this effect was reversed by the miR-134-3p inhibitors (Fig. 4C-E). These results suggested that PDIA6 was a direct target of miR-143-3p, and the expression levels of PDIA6 were positively modulated by PCAT6.
Figure 4.
PCAT6 upregulates the expression levels of PDIA6 via miR-143-3p. (A) The binding sites of miR-143-3p in 3'-untranslated region of PDIA6 were predicted by bioinformatics analysis. (B) Luciferase reporter assays were performed following co-transfection of the cells with WT-PDIA6 or MUT-PDIA6 and miR-143-3p mimics or NC mimics. (C-E) The (C) mRNA and (D and E) protein levels of PDIA6 were examined following transfection with siPCAT6 and anti-miR143. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05. miR, microRNA; si, small interfering RNA; NC, negative control; PCAT6, prostate cancer-associated transcript 6. WT, wild-type; MUT, mutant; PDIA6, protein disulfide isomerase A6.
Inhibition of miR-143-3p reverses the siPCAT6-mediated suppressive effects on the malignant phenotypes of bladder cancer cells
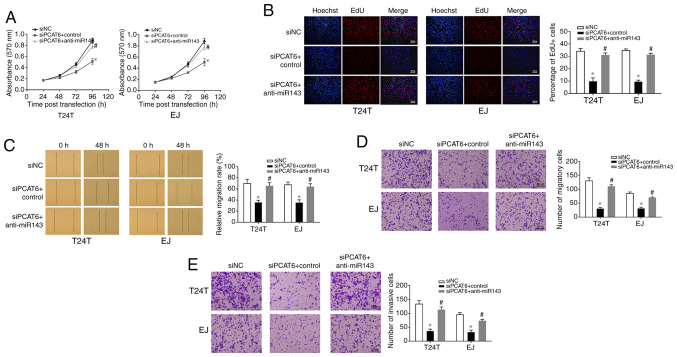
In order to determine whether PCAT6 affected bladder cancer cells via miR-143-3p, T24T and EJ cells were co-transfected with the miR-143-3p inhibitor and siPCAT6. As demonstrated in Fig. 5A and B, knockdown of PCAT6 led to a significant inhibition of cell proliferation compared with that in the siNC-transfected cells, whereas co-transfection with miR-143-3p inhibitor reversed this effect. Similarly, the results of the wound healing and Transwell assays revealed that the inhibition of miR-143-3p reduced the inhibitory effect of PCAT6 knockdown on cell migration and invasion (Fig. 5C-E). These results suggested that PCAT6 was effective in promoting the malignant behaviors of bladder cancer cells in a miR-143-3p dependent manner.
Figure 5.
miR-143-3p inhibition reverses the effects of PCAT6 depletion on cell proliferation, migration and invasion. (A and B) Cell proliferation was determined by (A) MTT and (B) EdU assays following transfection with siNC, siPCAT6 or siPCAT6 + anti-miR143. Scale bar, 100 µm. (C) Wound healing and (D) Transwell assays were performed to examine the effects of anti-miR143 on cell migration inhibition induced by siPCAT6. Scale bar, 100 µm. (E) Cell invasive ability was analyzed by Transwell assay with Matrigel following miR-143-3p inhibition. Scale bar, 100 µm. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 vs. siNC; #P<0.05 vs. siPCAT6 + control. miR, microRNA; si, small interfering RNA; NC, negative control; PCAT6, prostate cancer-associated transcript 6.
Discussion
Bladder cancer is one of the most common urinary malignancies worldwide with high morbidity and mortality rates (23). Recent studies have suggested that lncRNAs may serve essential roles in the development and malignant progression of bladder cancer (24,25). Therefore, the study of bladder cancer-related lncRNAs may help identify early therapeutic targets. lncRNA PCAT6 is a lncRNA that has been reported to be upregulated in osteosarcoma and colorectal cancer (26,27) and has emerged as a critical regulator in tumorigenesis and cancer progression. For example, Bai et al (28) have demonstrated that PCAT6 promotes gastrointestinal stromal tumor cell proliferation and stemness, but inhibits apoptosis through the ceRNA mechanism and the activation of the Wnt signaling pathway. Ma et al (14) have reported that PCAT6 facilitates the proliferation, migration and cisplatin resistance of cervical cancer cells and impedes their apoptosis via the PCAT6/miR-543/zinc finger E-box-binding homeobox 1 axis. However, the understanding of the effects of PCAT6 on bladder cancer carcinogenesis is limited. In the present study, the expression levels of PCAT6 were enhanced in bladder cancer tissues compared with that in adjacent normal tissues. The knockdown of PCAT6 suppressed bladder cancer cell proliferation, migration and invasion. According to these results, PCAT6 may be a suitable putative diagnostic biomarker and therapeutic target for bladder cancer. However, further studies are required to determine whether PCAT6 may regulate other biological behaviors of bladder cancer, such as apoptosis and chemoresistance.
To date, the ceRNA hypothesis has been extensively studied, and there increasing studies have analyzed the involvement of a complex network of lncRNA/miRNA/mRNA pathways in various types of human cancer, including bladder cancer (29-31). In the present study, miR-143-3p was demonstrated to be a direct target of PCAT6 in bladder cancer. PDIA6 belongs to the protein disulfide isomerase (PDI) family and is widely expressed in various types of human cancer, including breast cancer (32) and ovarian cancer (33). Increasing evidence suggests that PDIA6 is associated with the progression of various types of cancer (34-35). For example, Kim et al (36) reported that inhibition of PDIA6 may transduce EGFR signaling by activating and inducing A disintegrin and metalloprotease 17 during the migration and invasion of U87MG glioblastoma cells. Bai et al (37) have reported that PDIA6 is upregulated in NSCLC and inhibits cisplatin-induced NSCLC cell apoptosis and autophagy via the MAP4K1/JNK/c-Jun signaling pathway. In addition, PDIA6 has been demonstrated to be upregulated in bladder cancer, and its knockdown inhibits cell proliferation and invasion via the Wnt/b-catenin signaling pathway (38). In the present study, the expression levels of PDIA6 were suppressed by PCAT6 silencing in bladder cancer cells. Furthermore, rescue experiments revealed that miR-143-3p knockdown reduced the anticancer effects of PCAT6 silencing on cell proliferation, migration and invasion, suggesting that miR-143-3p/PDIA6 axis may be crucial for the tumorigenic roles of PCAT6 in bladder cancer.
In conclusion, the results of the present study revealed that PCAT6 downregulation inhibited cell proliferation, migration and invasion by targeting the miR-134-3p/PDIA6 axis. These results may provide a new theoretical basis to explore the mechanism underlying the regulation of bladder cancer and a novel target for its diagnosis and treatment.
Acknowledgements
Not applicable.
Funding Statement
Funding: This study was supported by The National Natural Science Foundation of China (grant no. 81802538) and The Health and Family Planning Commission of Wuhan (grant no. S201802260064).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GL designed the study, wrote and revised the manuscript. YZ and LC performed the experiments. YZ analyzed the data. GL, YZ and LC confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Central Hospital of Wuhan (2021-011) and written informed consent was obtained from each patient before enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Dettlaff K, Stawny M, Ogrodowczyk M, Jelińska A, Bednarski W, Wątróbska-Świetlikowska D, Keck RW, Khan OA, Mostafa IH, Jankun J. Formulation and characterization of EGCG for the treatment of superficial bladder cancer. Int J Mol Med. 2017;40:329–336. doi: 10.3892/ijmm.2017.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babjuk M, Bohle A, Burger M, Capoun O, Cohen D, Compérat EM, Hernández V, Kaasinen E, Palou J, Rouprêt M, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2016. Eur Urol. 2017;71:447–461. doi: 10.1016/j.eururo.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 4.Clark PE, Spiess PE, Agarwal N, Bangs R, Boorjian SA, Buyyounouski MK, Efstathiou JA, Flaig TW, Friedlander T, Greenberg RE, et al. NCCN guidelines insights: Bladder cancer, version 2.2016. J Natl Compr Canc Netw. 2016;14:1213–1224. doi: 10.6004/jnccn.2016.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohammed AA, El-Tanni H, El-Khatib HM, Mirza AA, Mirza AA, Alturaifi TH. Urinary bladder cancer: Biomarkers and target therapy, new era for more attention. Oncol Rev. 2016;10(320) doi: 10.4081/oncol.2016.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babjuk M. Bladder cancer in the elderly. Eur Urol. 2018;73:51–52. doi: 10.1016/j.eururo.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isin M, Dalay N. LncRNAs and neoplasia. Clin Chim Acta. 2015;444:280–288. doi: 10.1016/j.cca.2015.02.046. [DOI] [PubMed] [Google Scholar]
- 9.Wang M, Guo C, Wang L, Luo G, Huang C, Li Y, Liu D, Zeng F, Jiang G, Xiao X. Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell Death Dis. 2018;9(238) doi: 10.1038/s41419-018-0264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X, Wang M, Huang C, Wang L, Zeng F, Jiang G. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–291. doi: 10.1016/j.canlet.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi R, Wu P, Liu M, Chen B, Cong L. Knockdown of lncRNA PCAT6 Enhances radiosensitivity in triple-negative breast cancer cells by regulating miR-185-5p/TPD52 axis. Onco Targets Ther. 2020;13:3025–3037. doi: 10.2147/OTT.S237559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong D, Lun Y, Sun B, Sun H, Wang Q, Yuan G, Quan J. Silencing of long non-coding RNA PCAT6 restrains gastric cancer cell proliferation and epithelial-mesenchymal transition by targeting microRNA-15a. Gen Physiol Biophys. 2020;39:1–12. doi: 10.4149/gpb_2019044. [DOI] [PubMed] [Google Scholar]
- 14.Ma Z, Gu G, Pan W, Chen X. LncRNA PCAT6 accelerates the progression and chemoresistance of cervical cancer through up-regulating ZEB1 by sponging miR-543. Onco Targets Ther. 2020;13:1159–1170. doi: 10.2147/OTT.S232354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto H, Miller JS, Fajardo DA, Lee TK, Netto GJ, Epstein JI. Non-invasive papillary urothelial neoplasms: The 2004 WHO/ISUP classification system. Pathol Int. 2010;60:1–8. doi: 10.1111/j.1440-1827.2009.02477.x. [DOI] [PubMed] [Google Scholar]
- 16.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng F, Wang M, Li Y, Huang C, Tao D, Xie F, Zhang H, Sun J, Zhang C, Gu C, et al. CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett. 2019;460:139–151. doi: 10.1016/j.canlet.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Zhang H, Xie F, Tao D, Xiao X, Huang C, Wang M, Gu C, Zhang X, Jiang G. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2020;39:1696–1709. doi: 10.1038/s41388-019-1092-z. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang G, Wu AD, Huang C, Gu J, Zhang L, Huang H, Liao X, Li J, Zhang D, Zeng X, et al. Isorhapontigenin (ISO) inhibits invasive bladder cancer formation in vivo and human bladder cancer invasion in vitro by targeting STAT1/FOXO1 axis. Cancer Prev Res (Phila) 2016;9:567–580. doi: 10.1158/1940-6207.CAPR-15-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlevaro-Fita J, Johnson R. Global positioning system: Understanding long noncoding RNAs through subcellular localization. Mol Cell. 2019;73:869–883. doi: 10.1016/j.molcel.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmström PU, Choi W, Guo CC, Lotan Y, Kassouf W. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 24.Guo Y, Chen D, Su X, Chen J, Li Y. The lncRNA ELF3-AS1 promotes bladder cancer progression by interaction with Kruppel-like factor 8. Biochem Biophys Res Commun. 2019;508:762–768. doi: 10.1016/j.bbrc.2018.11.183. [DOI] [PubMed] [Google Scholar]
- 25.Wang F, Zu Y, Zhu S, Yang Y, Huang W, Xie H, Li G. Long noncoding RNA MAGI2-AS3 regulates CCDC19 expression by sponging miR-15b-5p and suppresses bladder cancer progression. Biochem Biophys Res Commun. 2018;507:231–235. doi: 10.1016/j.bbrc.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Guan X, Xu Y, Zheng J. Long non-coding RNA PCAT6 promotes the development of osteosarcoma by increasing MDM2 expression. Oncol Rep. 2020;44:2465–2474. doi: 10.3892/or.2020.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Zou Q, He H, Liang Y, Lei M, Zhou Q, Fan D, Shen L. Long non-coding RNA PCAT6 targets miR-204 to modulate the chemoresistance of colorectal cancer cells to 5-fluorouracil-based treatment through HMGA2 signaling. Cancer Med. 2019;8:2484–2495. doi: 10.1002/cam4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai F, Zhang N, Fang W, He X, Zheng Y, Gu D. PCAT6 mediates cellular biological functions in gastrointestinal stromal tumor via upregulation of PRDX5 and activation of Wnt pathway. Mol Carcinog. 2020;59:661–669. doi: 10.1002/mc.23199. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Wang ZL, Han CL, Wang MW, Jin Y, Jin XB, Xia QH. LncRNA CASC15 functions as an oncogene by sponging miR-130b-3p in bladder cancer. Eur Rev Med Pharmacol Sci. 2019;23:9814–9820. doi: 10.26355/eurrev_201911_19544. [DOI] [PubMed] [Google Scholar]
- 30.Zhou K, Yang J, Li X, Chen W. Long non-coding RNA XIST promotes cell proliferation and migration through targeting miR-133a in bladder cancer. Exp Ther Med. 2019;18:3475–3483. doi: 10.3892/etm.2019.7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Zu Y, Huang W, Chen H, Xie H, Yang Y. LncRNA CALML3-AS1 promotes tumorigenesis of bladder cancer via regulating ZBTB2 by suppression of microRNA-4316. Biochem Biophys Res Commun. 2018;504:171–176. doi: 10.1016/j.bbrc.2018.08.150. [DOI] [PubMed] [Google Scholar]
- 32.Ramos FS, Serino LT, Carvalho CM, Lima RS, Urban CA, Cavalli IJ, Ribeiro EM. PDIA3 and PDIA6 gene expression as an aggressiveness marker in primary ductal breast cancer. Genet Mol Res. 2015;14:6960–6967. doi: 10.4238/2015.June.26.4. [DOI] [PubMed] [Google Scholar]
- 33.Samanta S, Tamura S, Dubeau L, Mhawech-Fauceglia P, Miyagi Y, Kato H, Lieberman R, Buckanovich RJ, Lin YG, Neamati N. Expression of protein disulfide isomerase family members correlates with tumor progression and patient survival in ovarian cancer. Oncotarget. 2017;8:103543–103556. doi: 10.18632/oncotarget.21569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao L, Wu X, Gong Z, Yu M, Huang Z. PDIA6 contributes to aerobic glycolysis and cancer progression in oral squamous cell carcinoma. World J Surg Oncol. 2021;19(88) doi: 10.1186/s12957-021-02190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan C, Song X, Wang S, Wang J, Li L. Knockdown of PDIA6 inhibits cell proliferation and enhances the chemosensitivity in gastric cancer cells. Cancer Manag Res. 2020;12:11051–11062. doi: 10.2147/CMAR.S267711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim TW, Ryu HH, Li SY, Li CH, Lim SH, Jang WY, Jung S. PDIA6 regulation of ADAM17 shedding activity and EGFR-mediated migration and invasion of glioblastoma cells. J Neurosurg. 2017;126:1829–1838. doi: 10.3171/2016.5.JNS152831. [DOI] [PubMed] [Google Scholar]
- 37.Bai Y, Liu X, Qi X, Liu X, Peng F, Li H, Fu H, Pei S, Chen L, Chi X, et al. PDIA6 modulates apoptosis and autophagy of non-small cell lung cancer cells via the MAP4K1/JNK signaling pathway. EBioMedicine. 2019;42:311–325. doi: 10.1016/j.ebiom.2019.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng HP, Liu Q, Li Y, Li XD, Zhu CY. The inhibitory effect of PDIA6 downregulation on bladder cancer cell proliferation and invasion. Oncol Res. 2017;25:587–593. doi: 10.3727/096504016X14761811155298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.