Figure 5. Guided, programmable nucleases for ABDx.
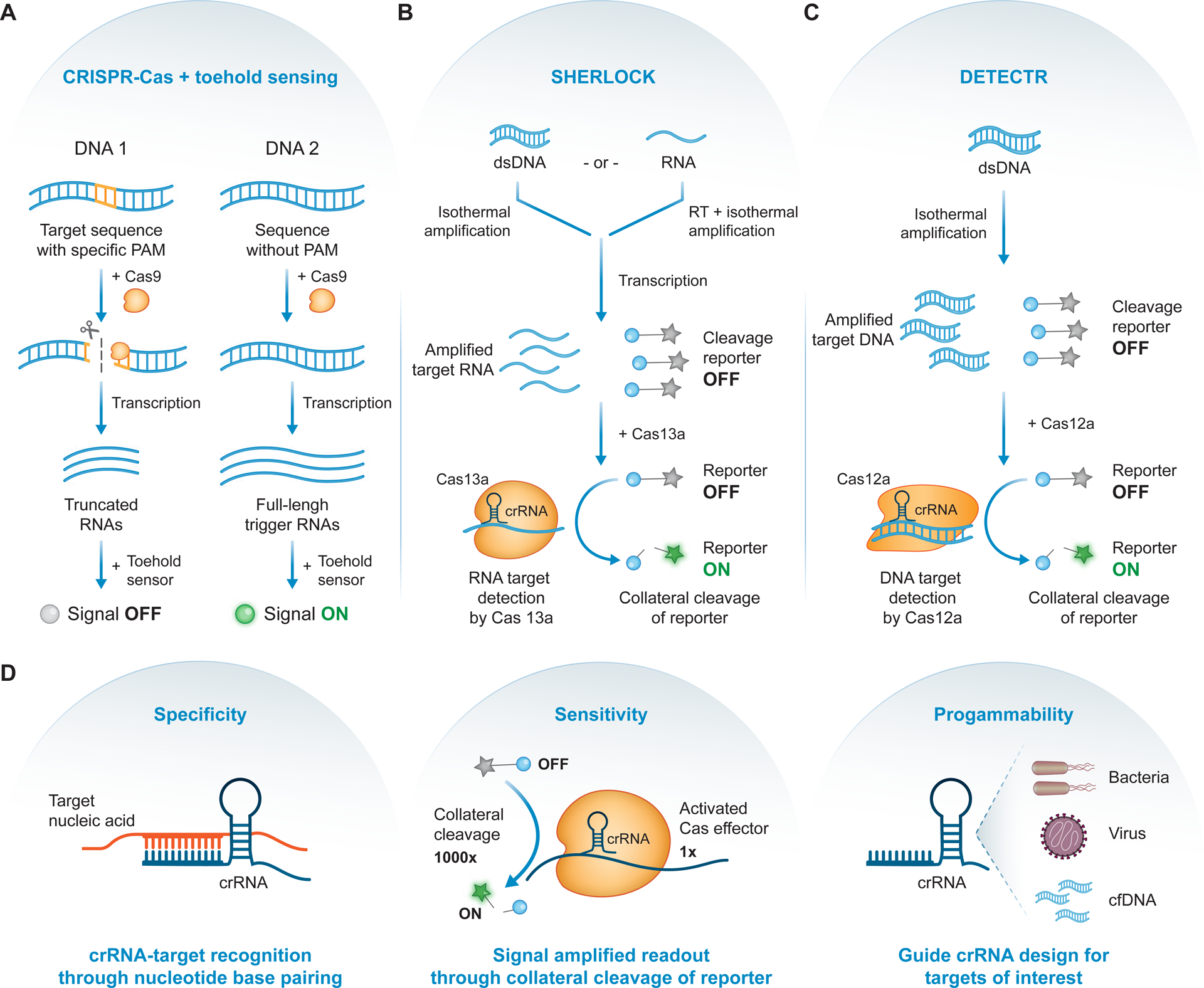
(A) Nucleic acid detection with CRISPR-Cas augmented toehold sensing. The presence of a specific protospacer adjacent motif (PAM; orange) in a DNA input results in cleavage by Cas9 with a PAM-specific guide RNA. Following transcription, this differential cleavage leads to either truncated or full-length trigger RNAs, which can be designed to differentially activate an engineered toehold sensor. Since only full-length triggers can activate the toehold sensor, a downstream signal can be used to discriminate the two inputs [111]. (B) SHERLOCK diagnostic test. Nucleic acids, either double-stranded DNA (dsDNA) or RNA, are amplified or reverse-transcribed (RT) and amplified, for DNA or RNA respectively. Following transcription, amplified target RNAs and fluorescently quenched cleavage reporters (grey star = fluorophore, blue circle = quencher) are incubated with Cas13a-crRNA complex. Detection and binding of the RNA target unleashes Cas13a’s collateral activity, resulting in reporter cleavage, fluorophore activation (green star), and signal amplification [114]. (C) DETECTR diagnostic test. Target dsDNA are amplified and, along with fluorescently quenched cleavage reporters, incubated with Cas12a-crRNA complex. Detection and binding of the dsDNA target unleashes Cas12a’s collateral activity, resulting in reporter cleavage, fluorophore activation, and signal amplification [115]. (D) CRISPR diagnostics achieve high specificity driven by crRNA guide-target base pairing (left), detection sensitivity from the highly efficient collateral cleavage of reporter molecules (middle), and diagnostic programmability due to facile guide RNA design for nucleic acid targets of interest (right).
