Abstract
Background
Pichia pastoris (syn. Komagataella phaffii) is an important yeast system for heterologous protein expression. A robust P. pastoris mutant with oxidative and thermal stress cross-tolerance was acquired in our previous study. The robust mutant can express a 2.5-fold higher level of lipase than its wild type (WT) under methanol induction conditions.
Results
In this study, we found that the robust mutant not only can express a high level of lipase, but also can express a high level of other heterogeneous proteins (e.g., green fluorescence protein) under methanol induction conditions. Additionally, the intracellular reactive oxygen species (ROS) levels in the robust mutant were lower than that in the WT under methanol induction conditions. To figure out the difference of cellular response to methanol between the WT and the robust mutant, RNA-seq was detected and compared. The results of RNA-seq showed that the expression levels of genes related to antioxidant, MAPK pathway, ergosterol synthesis pathway, transcription factors, and the peroxisome pathway were upregulated in the robust mutant compared to the WT. The upregulation of these key pathways can improve the oxidative stress tolerance of strains and efficiently eliminate cellular ROS. Hence, we inferred that the high heterologous protein expression efficiency in the robust mutant may be due to its enhanced oxidative stress tolerance. Promisingly, we have indeed increased the expression level of lipase up to 1.6-fold by overexpressing antioxidant genes in P. pastoris.
Conclusions
This study demonstrated the impact of methanol on the expression levels of genes in P. pastoris and emphasized the contribution of oxidative stress tolerance on heterologous protein expression in P. pastoris. Our results shed light on the understanding of protein expression mechanism in P. pastoris and provided an idea for the rational construction of robust yeast with high expression ability.
Keywords: Yeast, Pichia pastoris, Heterologous protein expression, Lipase, Oxidative stress tolerance
Background
With its proven ability to express over 400 proteins, from human endostatin to spider dragline silk protein, Pichia pastoris has become a consistent choice for heterologous protein production [1–3]. Additionally, P. pastoris as a eukaryotic organism is capable of secreting high titres of correctly folded, post-translationally processed and active recombinant proteins into the culture media [4, 5]. P. pastoris has the ability to produce proteins of therapeutic and commercial interest in concentrations ranging from milligrams to grams per litre [6]. Fermentations can be readily scaled up to meet greater demands, and factors influencing protein productivity and activity, can be controlled. Furthermore, as P. pastoris does not secrete high levels of native proteins, the purification of secreted makings is much easier than in other systems [7].
Many methods have been used to improve the expression of heterologous proteins in P. pastoris, mainly by codon optimization, increasing gene dosage, promoting protein folding and secretion, etc. Codon usage differs among organisms, thereby substituting rare codons basing on the preferred codon usage of P. pastoris is vital to realize high expression levels of heterologous proteins. The efficient production of a novel pH-stable xylanase from the fungus Corynascus thermophilus was achieved by codon optimization [8]. Simply increasing the copy number of target genes can improve the production of target heterologous proteins [9, 10]. However, the overexpression of heterologous protein might cause the accumulation of unfolded and misfolded protein in the endoplasmic reticulum (ER), activating the unfolded protein response (UPR) [11]. After that, the ER-associated degradation pathway can be activated and then reduce the production of heterologous proteins. Based on the above rationales, to prevent the degradation of overexpressed heterologous protein by the proteasome, the disruption of Pep4 protease increases phytase secretion in P. pastoris [12]. Lipase is widely used in the production of biodiesels [13], detergents [14], chiral compounds [15], etc. Protein disulfide isomerase (PDI), a chaperone in ER, can catalyse the disulfide bond formation and help in correcting the folding of a protein. Previously, we successfully improved the expression level of lipase in P. pastoris by co-expressing PDI [16].
During fermentation process, cells may encounter multiple environmental stresses from the fermentation medium, product, temperature, etc. These environmental stresses not only reduce the production of products, but even limit the growth of yeast cells. As we know, H2O2 generated by the metabolism of methanol may cause oxidative stress and impact the production efficiency of P. pastoris. However, research had barely reported the effect caused by methanol on the production of heterologous protein and the change of cellular response in P. pastoris. Additionally, few researchers have tried to strengthen the heterologous protein expression ability in P. pastoris from the perspective of improving their oxidative stress tolerance. Indeed, many other researchers have successfully increased the production efficiency of yeast strains by relieving repression and improving their stress tolerance. These examples provide a good reference for the modification in P. pastoris. For example, researchers have greatly improved the ethanol production of a Kluyveromyces marxianus mutant by adaptive evolution in 6% (v/v) ethanol [17]. The glucose and xylose fermentation in Saccharomyces cerevisiae under acetic acid stress conditions was improved by overexpressing RCK1 coding for a protein kinase involved in oxidative stress [18].
Previously, a P. pastoris mutant with thermal and oxidative stress cross-tolerance was found to have a higher lipase expression ability than its WT under methanol induction conditions. In this study, we found that this phenomenon was also present in the expression of other protein. Given the high oxidative stress tolerance in the robust mutant, we examined the changes of the intracellular reactive oxygen species (ROS) levels in the WT and the robust mutant under methanol-induced conditions. Additionally, the transcriptomes of the WT and the robust mutant during the methanol-induced fermentation process and growth condition were also explored and compared. As a result, we found that the mechanism of high heterologous protein expression ability in the robust mutant was mainly owed to its enhanced oxidative stress tolerance. To validate this inference, we rationally designed antioxidant gene-overexpressed strains, and successfully improved the heterologous protein expression ability in these strains compared to their blank control. This study emphasized the importance of oxidative stress tolerance in protein expression in P. pastoris and provided ideas for rationally increasing the production efficiency of heterologous protein in the future.
Results
Improved heterologous protein expression ability in the robust mutant under methanol induction
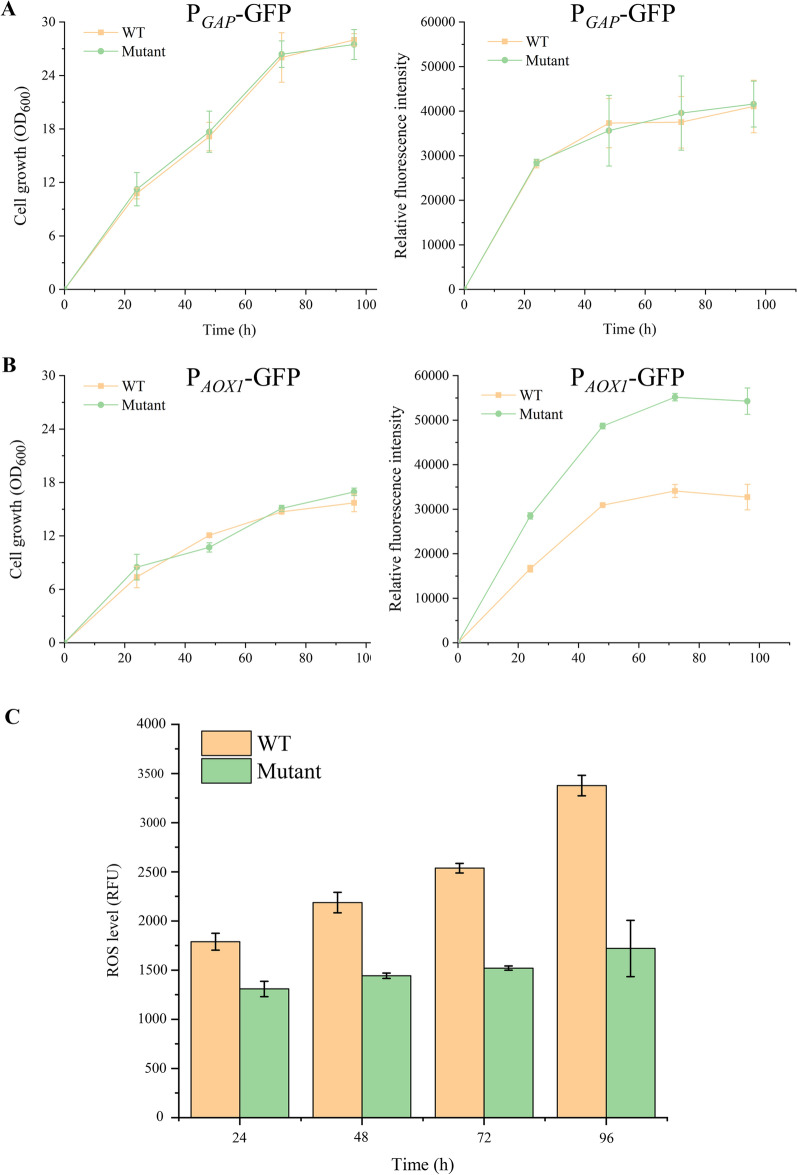
Previously, we isolated a robust P. pastoris mutant, which exhibited thermal and oxidative co-stress tolerance. In addition, the robust mutant expressed an up to 2.5-fold higher level of heterologous protein than the WT [19]. To determine whether this phenomenon could also occur in the expression of other proteins, a green fluorescence protein (GFP) was expressed in the robust mutant and its WT. Two promoters, PGAP and PAOX1, were used to initiate the expression of GFP. The alcohol oxidase 1 (AOX1) promoter (PAOX1) of P. pastoris is one of the most widely used promoters for expressing a large number of proteins. The action of PAOX1 is intensely repressed by multiple carbon sources such as glucose and glycerol, but is strongly induced by methanol [20]. PGAP is a common strong constitutive promoter from the glyceraldehyde 3-phosphate dehydrogenase gene (GAP) [21]. Under the control of PGAP, the expression levels of GFP were similar in the WT and the robust mutant (Fig. 1A). However, under the control of PAOX1 with methanol as an inducer, the expression level of GFP in the robust mutant was 1.3-fold higher than that of in the WT (Fig. 1B). In addition, the cell growth curve showed that the growth of the WT and the robust mutant under methanol condition was inhibited compared with that under glucose condition. The results of GFP expression in the WT and the robust mutant indicated that the robust mutant expressed a higher level of heterologous proteins under methanol induction. We inferred that the robust mutant might be more capable to overcome the oxidative stress caused by methanol than its WT, leading to its enhanced protein expression ability. We then detected the intracellular ROS levels of the robust mutant and the WT and tried to analyse the expression mechanism of the robust mutant and the WT by RNA-seq.
Fig. 1.
The characteristics of the WT and the robust mutant. A The cell growth and fluorescence intensity of green fluorescence protein (GFP) in the WT and the robust mutant under glucose condition; B the cell growth and fluorescence intensity of GFP in the WT and the robust mutant under methanol condition; C ROS levels of the WT and the robust mutant during fermentation process under the induction of methanol
Enhanced anti-oxidative stress ability helps to reduce the accumulation of ROS in P. pastoris
Under the control of PAOX1, methanol is used as an inducer and a sole carbon resource for fermentation with P. pastoris. The metabolism of methanol generates hydrogen peroxide (H2O2), which is a kind of ROS [22]. During the fermentation process under the induction of methanol, the intracellular ROS levels of WT and the robust mutant were measured and compared (Fig. 1C). The accumulation of ROS in cells gradually increased over time. However, the accumulations of ROS in the WT were higher up to twofold than that in the robust mutant. The result indicated that a stronger ROS buffering system may be in the robust mutant to efficiently reduce the ROS level caused by methanol. Hence, RNA-seq of the robust mutant and the WT was carried out to further explore the underlying mechanism.
Augmented ROS buffering capacity renders higher expression ability based on transcriptome analysis
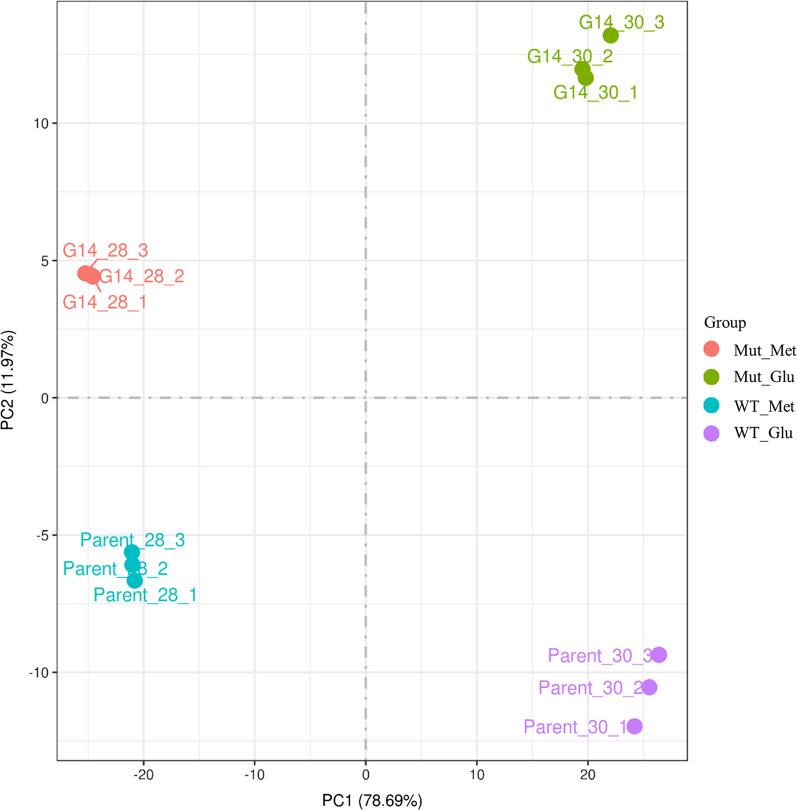
The heterologous protein expression ability of P. pastoris can mainly be mediated by cellular stress, transcription, protein folding, secretion pathway, and carbon metabolism pathway. To explore the underlying mechanism, the transcriptomes of the WT and the robust mutant were analysed and compared under methanol-induced conditions. Yeast cells were collected at 60 h for RNA-seq, as enzyme activity of these P. pastoris strains sharply increased at 60 h. The data of RNA-seq was been uploaded in NCBI’s Gene Expression Omnibus (GEO) public archive database and the Accession number is GSE167141. As shown in Fig. 2, the principal-component analysis (PCA) of transcriptome data showed that the triplicates of each sample were similar, indicating that the data could be employed for the following analysis.
Fig. 2.
The correlation of inter-group samples by PCA analysis. Three biological replicates were used in each group
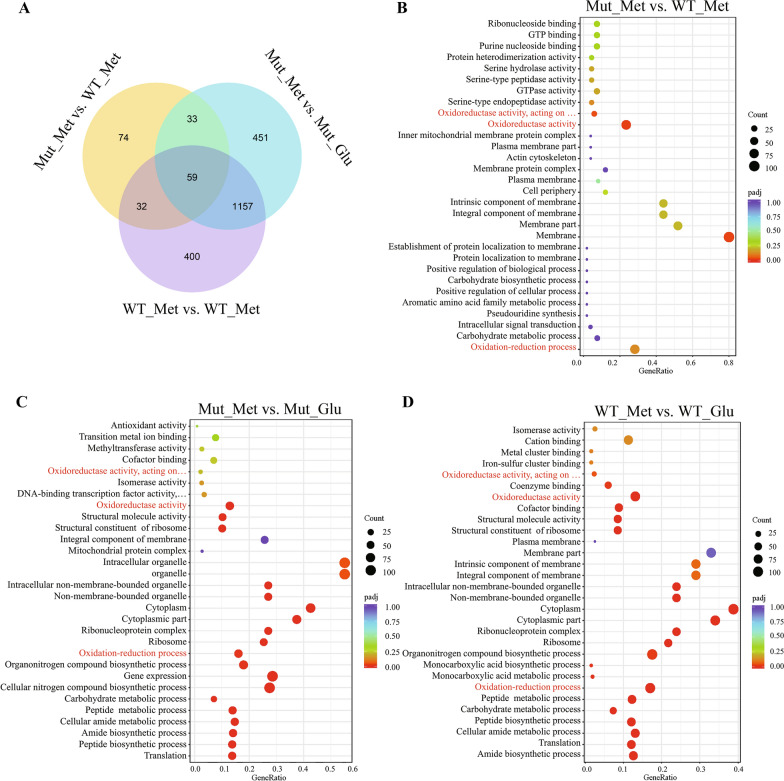
The analysis of differentially expressed genes was focused on the WT and the robust mutant under methanol induction conditions and glucose culture conditions (previously uploaded in NCBI’s GEO, GSE157242). Three comparison groups were set to explore the cellular response to methanol in P. pastoris strains (Fig. 3). The comparison group Mut_Met versus WT_Met represented the robust mutant under methanol condition versus the WT under methanol condition. The comparison group Mut_Met versus Mut_Glu represented the robust mutant under methanol condition versus that in glucose condition. The comparison group WT_Met versus WT_Glu represented the WT under methanol condition versus that in glucose condition. With the absolute value of log2-fold change > 1 and p-value < 0.05 as classification criteria, the number of differentially expressed genes of each comparison group are listed Venn diagram (Fig. 3A). In the comparison group Mut_Met versus WT_Met, 198 genes were differentially expressed. Methanol caused stress on the WT and the robust mutant, so that a large number of differentially expressed genes were found in the other two comparison groups. A Gene Ontology (GO) enrichment analysis of differentially expressed genes in each comparison group was performed to identify enriched GO terms, including biological process (BP), cellular component (CC), and molecular function (MF). Interestingly, the biological process of oxidation–reduction and the molecular function of oxidoreductase activity were enriched in all these three comparison groups (Fig. 3B–D). The result demonstrated that the oxidative stress caused by methanol may lead to active intracellular redox changes. Moreover, the robust mutant has an enhanced oxidative stress tolerance. The enrichment of GO terms related to redox in comparison group Mut_Met versus WT_Met may explain the reduced ROS levels in the robust mutant.
Fig. 3.
Transcriptomic analysis. A The Venn diagram of differentially expressed genes in three comparison groups. B–D Bubble diagrams of GO enrichment result, each ten GO terms from bottom to top represents biological process, cellular component, and molecular function, respectively. B GO enriched analysis of comparison group Mut_Met vs. WT_Met; C GO enriched analysis of comparison group Mut_Met vs. Mut_Glu; D GO enriched analysis of comparison group WT_Met vs. WT_Glu. The GO terms marked in red include oxidation–reduction process (GO:0055114), oxidoreductase activity (GO:0016491), oxidoreductase activity, acting on CH–OH group of donors (GO:0016614), and oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen (GO:0016705)
The factors related to redox mainly include MAPK pathway, peroxisome, stress response transcription factors, antioxidant defence system, and ergosterol synthesis pathway, so we then investigated the expression levels of related genes (Fig. 4). The MAPK pathway is responsible for resisting environmental stresses [23]. Interestingly, the MAPK pathway (Fig. 4A) was upregulated in all three comparison groups, further indicating methanol is a kind of stress to P. pastoris. Furthermore, the upregulation phenomenon was obvious in the comparison group Mut_Met versus WT_Met, indicating that the environmental stress response in the robust mutant was stronger than that in the WT.
Fig. 4.

Heat map profiles of key genes. A Genes related to MAPK pathway; B genes related to peroxisome; C genes related to antioxidants and transcription factors responded to oxidative stress; D the ergosterol synthesis pathway
The peroxisome pathway (Fig. 4B) was tremendously upregulated in the comparison group Mut_Met versus Mut_Glu and WT_Met versus WT_Glu as the first step of methanol metabolism is in peroxisomes [24]. Peroxisomes possess enzymes that degrade ROS, especially catalase [25]. Predictably, the expression levels of these peroxisomal protein encoding genes were also considerably upregulated in the robust mutant versus the WT under identical methanol conditions, explaining the higher protein expression level in the robust mutant.
Because of the stressful condition caused by methanol, the expression levels of two oxidative stress transcription factors, YAP1 and SKN7, were upregulated in these three comparison groups. The expression levels of several genes related to ROS scavengers (Fig. 4C) were tremendously upregulated in all comparison groups, especially CAT (encoding catalase) and PMP20 (encoding a glutathione peroxidase). Catalase is responsible for removing H2O2 [26], while PMP20 is contributed to the removal of other oxidized molecules [27]. The results indicated that more ROS need to be eliminated in the WT and the robust mutant under methanol condition than that in glucose condition. Interestingly, the ergosterol synthesis pathway (Fig. 4D) was showing upregulation in these comparison groups. Other researchers [28] proved that the addition of ergosterol can repress the production of ROS by the NADPH oxidase (NOX). Interestingly, from the result of RNA-seq, the expression level of NOX in the robust mutant is 15% lower than that in the WT. Consistently, most genes related to the ergosterol synthesis pathway in comparison group Mut_Met versus WT_Met were upregulated, further explaining the reduced ROS level in the robust mutant.
The upregulation of Mxr1 and Prm1 improves the efficiency of the AOX1 promoter
PAOX1 is commonly used to initiate the transcription of heterologous proteins in P. pastoris, and its transcription efficiency is mediated by transcription factors (TFs) (Fig. 5). Binding sites for TFs, methanol-induced transcription factor 1 (encoded by Mit1) [29, 30], methanol expression regulator (encoded by Mxr1) [31] and a positive transcription regulator for methanol-utilization genes (encoded by Prm1) [32], had been identified on the sequence of PAOX1. These TFs positively regulate the function of PAOX1. The transcription levels of Mit1, Mxr1, Prm1, and heterologous lipase gene (RCL) were measured and compared between the WT and the robust mutant. The expression levels of RCL, Mxr1, and Prm1 were markedly improved in comparison group Mut_Met versus WT_Met according to the results of RNA-Seq and RT-qPCR (Table 1). The expression level of RCL was up to 4.2-fold in the robust mutant than that in WT, explaining the higher lipase activity in the robust mutant. The expression level of transcription factors Mxr1 and Prm1 in the robust mutant was up to 1.6-fold and 1.8-fold higher than that in the WT, respectively. Therefore, the higher expression level of heterologous protein in the robust mutant may be partly due to the improved efficiency of PAOX1.
Fig. 5.
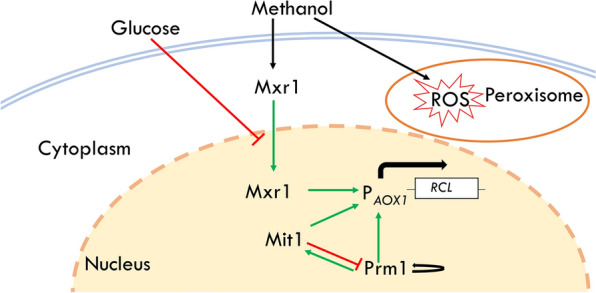
The predicted regulatory model of PAOX1 activation. Green arrows indicate activation; red blunt-end arrows indicate repression
Table 1.
The expression levels of genes related to the activation of PAOX1 by RNA-Seq and RT-qPCR
| Gene name | Foldchange (RNA-seq) | Adjusted p-value | Foldchange (RT-qPCR) | p-value | |
|---|---|---|---|---|---|
| RCL | 3.7 | *** | 4.2 | * | |
| Mxr1 | 1.2 | *** | 1.6 | ** | |
| Prm1 | 1.2 | *** | 1.8 | ** | |
| Mit1 | 0.98 | ns | 1.2 | ns | |
Results were means of three biological replicate experiments
ns not significant
*p < 0.05; **p < 0.01; ***p < 0.001
The downregulation of the methanol dissimilatory pathway improves the production of heterologous protein
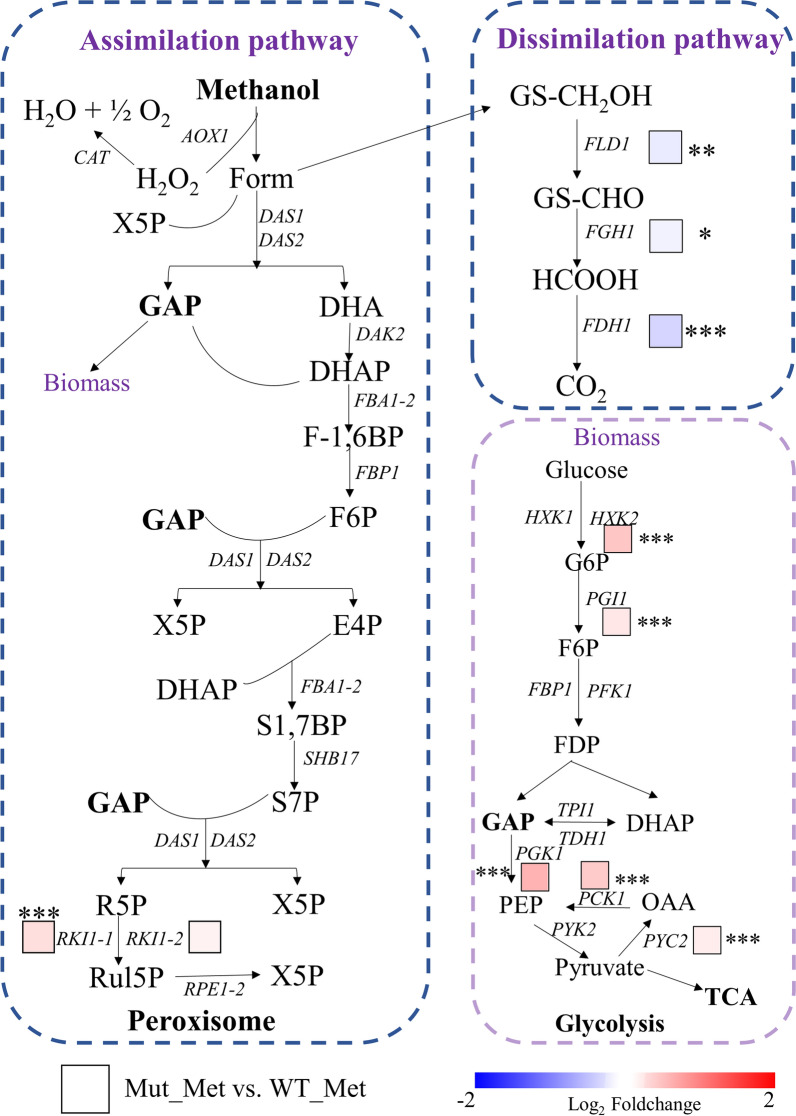
Pichia pastoris is a methylotrophic yeast, able to utilize methanol for energy production and to assimilate it as a sole carbon source for cell growth and producing target product [33]. The metabolism of methanol as part of carbon metabolism consists of two pathways, the assimilatory pathway and the dissimilatory pathway (Fig. 6). The assimilatory pathway of methanol is in peroxisomes of P. pastoris [24]. In the assimilatory pathway, methanol participates in the carbon metabolism to produce organic matters like endogenous and exogenous proteins. However, in the dissimilatory pathway, methanol is ultimately transformed into carbon dioxide. Mapping the results of RNA-seq into the methanol metabolism network, the dissimilatory pathway (FLD1, FGH1, and FDH1) of methanol was down-regulated in Mut_Met versus WT_Met, while some genes (RKI1-1 and RKI1-2) involving in the assimilatory pathway were upregulated. The methanol in the robust mutant was more assimilated to generate biomass that genes (HXK2, PGI1, PGK1, PCK1, and PYC2) involved in glycolysis were significantly upregulated in comparison group Mut_Met versus WT_Met. These results indicated that the efficiency of the methanol assimilatory pathway may be improved to generate more product, which also explained the higher protein expression level in the robust mutant.
Fig. 6.
Expression levels of key genes. Comparative of the robust mutant with the wild type under identical methanol conditions. AOX1: alcohol oxidase 1; Form, formaldehyde; DAS1, dihydroxyacetone synthase; DHA: dihydroxyacetone; DHAP: dihydroxyacetone phosphate; E4P: erythrose-4-phosphate; FBA1-2: fructose 1,6-bisphosphate aldolase; FDH1: formate dehydrogenase 1; Form, formaldehyde; F1,6BP: fructose 1,6-bisphosphate; F6P: fructose-6-phosphate; GAP: glyceraldehyde-3-phosphate; R5P: ribose-5-phosphate; Rul5P: ribulose-5-phosphate; S1,7BP: sedoheptulose-1,7-bisphosphate; S7P: sedoheptulose-7-phosphate; X5P: xylulose-5-phosphate; GS-CH2OH: S-(hydroxymethyl)glutathione; GS-CHO: S-formylglutathione; HCOOH: formic acid; G6P: glucose-6-phosphate; FDP: fructose-1, 6-diphosphate; PEP: phosphoenolpyruvate; OAA: oxaloacetic acid; TCA: tricarboxylic acid cycle. *p < 0.05, **p < 0.01, ***p < 0.001
Rational design to improve lipase expression level based on the overexpression of antioxidant genes
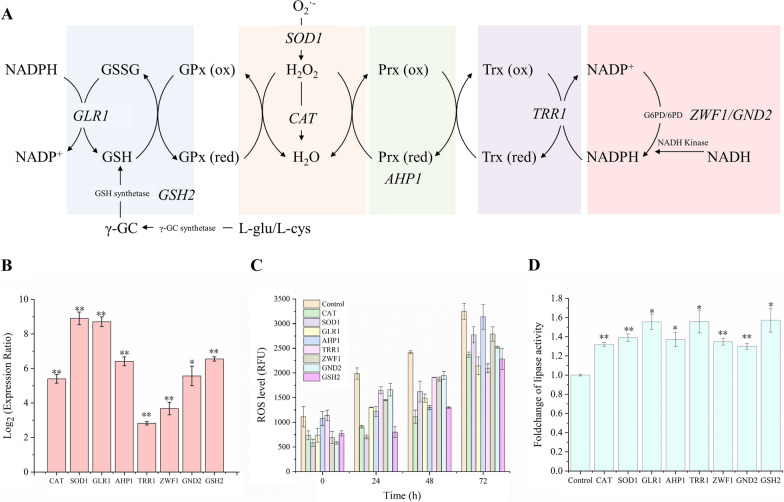
Based on the results above, we believed the engineering of the cell’s antioxidant defence system by overexpression of genes involved in endogenous antioxidants could alleviate the toxic effects induced by methanol, and thereby can increase the protein expression level in P. pastoris. According to the antioxidant defence system [34, 35] (Fig. 7A), CAT, SOD1, GLR1, AHP1, TRR1, ZWF1, GND2, and GSH2 was solely overexpressed by constitutive promoter PGAP in P. pastoris hosts. The SOD1 gene encoding superoxide dismutase 1 converts superoxide anion (O2·−) to H2O2. The CAT encoding catalase directly degrades H2O2. Glutathione is one of the main antioxidants in living cells [36]. The GLR1 encoding glutathione reductase helps to make reduced glutathione. The GSH2 encoding glutathione (GSH) increases total glutathione content. In yeast cells, thioredoxins partner with peroxiredoxin in H2O2 signalling [34]. The AHP1 encoding alkyl hydroperoxide reductase 1 is a member of the peroxiredoxin family to promote the elimination of cellular H2O2 [37]. The TRR1 encoding thioredoxin reductase plays an important role in thioredoxin-dependent antioxidant pathways [38]. The reducing power provided by NADPH is generated from the pentose-phosphate pathway via glucose-6-phosphate dehydrogenase (encoded by ZWF1) and 6-phosphogluconate dehydrogenase (encoded by GND2) [39]. The relative transcription levels of these genes in gene-overexpressing strains were measured by RT-qPCR. As shown in Fig. 7B, these genes were successfully overexpressed.
Fig. 7.
Overexpression of antioxidant genes to improve the heterologous protein expression ability of P. pastoris. A The antioxidant defence system of yeasts. O2·−: superoxide anion; SOD: superoxide dismutase; CAT: catalase; GPx: glutathione peroxidase; GLR1: glutathione reductase; Prx: peroxiredoxin/thioredoxin peroxidase; Trx: thioredoxin; TRR1: thioredoxin reductase; G6PD: glucose-6-phosphate dehydrogenase; 6PD: 6-phosphogluconate dehydrogenase; GSH: glutathione; γ-GC: gamma glutamylcysteine; l-glu: l-glutamic acid; l-cys: l-cysteine. B Relative transcription levels of antioxidant system-related genes. The expression ratio of a gene was analysed using the 2−ΔΔCt method; C the intracellular ROS level of overexpressed strains and their blank control during methanol-induced fermentation; D foldchange of lipase activity levels in overexpressed strains. *p < 0.05, **p < 0.01
To explore the contribution of antioxidant genes in lipase expression, flask-batch fermentation of each overexpressed strain was performed (Fig. 7C, D). Under the control of PAOX1, the lipase was expressed using methanol as an inducer. During the process of fermentation, the intracellular ROS levels of overexpression strains and the control were measured and compared. As shown in Fig. 7C, the ROS levels in these overexpressed strains were significantly lower than that of the blank control. It indicated that the contributions of antioxidant genes were very considerable. Compared to the blank control, the expression levels of lipase in overexpressed strains were improved up to 1.6-fold (Fig. 7D). These results revealed that overexpression of antioxidant genes improved the expression of lipase under methanol induction condition.
Discussion
As we know, H2O2 is generated as a by-product of methanol metabolism, causing oxidative stress on P. pastoris cells [40]. The accumulation of ROS reduced the production of proteins. However, the response mechanism of P. pastoris to methanol is still unclear. Previously, we obtained a robust P. pastoris mutant with a strong ability to express heterogeneous proteins, which could be used as a good start point for revealing the underlying mechanism responding to oxidative stress under methanol induction. It is interesting to explore the differences in cellular response between the strains with different protein expression abilities. After RNA-seq, we found the GO terms related to redox were enriched in the comparison groups under methanol induction conditions versus glucose conditions (Fig. 3). Moreover, under methanol induction conditions, GO terms related to redox were also enriched in the robust mutant versus the WT. The results revealed that the robust mutant exhibited a lower level of ROS and enhanced protein production efficiency owing to its augmented oxidative stress tolerance. In addition, we improved the methanol-induced lipase expression ability of non-oxidative stress-tolerant strain by rationally increasing its ROS buffering capacity, further explaining the importance of strains with strong oxidative stress tolerance in methanol-induced fermentation.
Compared to the WT, we found that the antioxidant property of the robust mutant was mainly attributed to four characteristics: active redox processes, active environmental stress response (upregulation of MAPK pathway), higher expression levels of antioxidant-related transcription factors, and upregulation of ergosterol synthesis pathways. The results supported again the conclusion that intracellular redox balance is very important for the cell productivity of yeast [41]. The imbalance of redox will cause the accumulation of intracellular ROS, leading to protein peroxidation [42] and lipid peroxidation [43]. These disorders reduce the production of products in yeast. Hence, maintaining redox homeostasis in hosts will efficiently enhance their productivity. The antioxidant defence system in yeasts mainly consists of the enzymatic part and non-enzymatic part [35]. The key parts of the antioxidant defence system are shown in Fig. 7A. Superoxide dismutase, catalase, and peroxiredoxins are the main members of the enzymatic defence system. The non-enzymatic defence system generally includes GSH, thioredoxin, vitamin C (ascorbic acid), and other reducing agents. In addition, cofactor engineering for enhancing the production of products and improving cell growth has become popular. In cells, the pentose phosphate pathway is activated to compensate for the severe request of NADPH upon oxidative stress [44]. Besides, augmenting NADPH levels promoted protein production in P. pastoris [45].
As reported previously, it is an efficient way to enhance cell productivity by the overexpression of antioxidant genes. The production of ethanol was boosted by overexpressing multilevel defence system which effectively reduced the accumulation of cellular ROS and alleviated lipid peroxidation in S. cerevisiae [46]. A robust lipid production platform of Yarrowia lipolytica was built by modulation of oxidative stress defence pathway [47]. In this study, the overexpression of genes related to antioxidant defence system reduced the cellular ROS level during methanol-induced fermentation and then improved the production of lipase (Fig. 7). The researchers also found that the combination of effective genes can sometimes double the effect [46]. Therefore, in the follow-up of this study, we consider overexpressing the combination of these functional genes in P. pastoris, looking forward to obtaining strains with stronger protein expression potential.
The methods to improve the oxidative stress tolerance of yeast cells are not limited to simply overexpressing the genes encoding antioxidant enzymes. It is also a good way to start by repressing the source of ROS generation. Promisingly, this study revealed that the ergosterol pathway was significantly upregulated in strains under methanol condition versus glucose condition and the comparison group Mut_Met versus WT_Met (Fig. 4D). The increase in ergosterol content is conducive to inhibiting the production of intracellular ROS by NOX [48]. And, the supplementation of ergosterol mitigated oxidative stress in S. cerevisiae [49]. The mutants lacking ergosterol are hypersensitive to oxidative stress [50]. Moreover, Caspeta et al. [51] have proved that the ergosterol pathway is importantly related to the thermotolerance of yeast. Sterols including ergosterol are the main components of cell membranes. Adjusting the ratio of cell membrane components can also enhance cell tolerance [52]. Hence, the ergosterol synthesis pathway will be our future target to rationally enhance the oxidative stress tolerance of P. pastoris.
Conclusions
This study found that a robust P. pastoris mutant with oxidative stress tolerance can express high levels of heterologous proteins under the induction of methanol. From the results of ROS levels and RNA-seq, we proved that methanol mainly induces the oxidative stress response in P. pastoris. Based on the importance of antioxidant defence system, we rationally improved the production of heterologous protein lipase up to 1.6-fold by overexpressing genes related to antioxidant defence system in P. pastoris. However, it is difficult to achieve the desired effect only by overexpression of a single functional gene. In the future, we will consider further improve the heterologous protein expression ability of P. pastoris from the view of global regulation. Our findings provide a further understanding of the impact of methanol on the expression of heterologous proteins in P. pastoris and highlight potential targets for constructing yeasts with high protein expression potential.
Methods
Strain and plasmid construction
The robust mutant was previously evolved by adaptive laboratory evolution [19]. Both the robust mutant and its WT expressed a recombinant protein lipase encoded by RCL gene and tightly regulated by the methanol-inducible promoter PAOX1. For gene overexpression, the antioxidant genes from the genome of P. pastoris were constructed into the pGAPZα vector using MultiF SeamLess Assembly Mix (ABclonal, China). The plasmids were introduced into P. pastoris-competent cells by electroporation, using an Eppendorf Eporator (Eppendorf, Germany).
Expression of heterologous proteins
Under the control of inducible promoter PAOX1, the fermentation process was performed in BMGY/BMMY fermentation medium containing 100 mM potassium phosphate (pH 6.0), 1.34% yeast nitrogen base without amino acids, 4 × 10−5% biotin, and 1% glycerol (BMGY) or 0.5% methanol (BMMY) at 28 °C [16]. Methanol as a sole carbon source and inducer was supplied into the medium every 24 h. The fermentation broth of each sample expressing lipase was centrifuged, and aliquots of the supernatant were diluted to measure the lipase activities. The lipase enzyme activity of samples was assayed according to Sha et al. [16]. Under the control of constitutive promoter PGAP, the fermentation process of GFP was conducted in YPD liquid medium. The fermentation broth of each sample expressing GFP was centrifuged, and cell pellets were resuspended to measure fluorescence intensity at 485/528 excitement/emission (nm). The fluorescence intensity was normalized to 1 OD600.
Measurement of ROS level
Intracellular ROS was measured using 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA) [53] by three biological replicates of each sample. Briefly, the cells were harvested by centrifugation, resuspended in PBS, and treated in 10 μM DCFH-DA dissolved in DMSO for 1 h. After centrifugation, the cell pellets were collected and resuspended in PBS. After crushing by glass bead disruption, the supernatant was collected, and DCF fluorescence intensity was measured at 480/525 excitement/emission (nm). The protein concentration was measured by the Bradford method [54]. DCF fluorescence intensity was normalized to the protein level of the supernatant.
RNA-sequencing
Samples for RNA-seq were collected from fermentation cultures at 60 h. Three biological replicates of each sample were adopted to ensure data reliability. The cDNA library construction and sequencing, quality control, and alignment to P. pastoris genome were provided by Novogene, Beijing, China (http://www.novogene.cn/). Genes with an adjusted p-value < 0.05 found by DESeq2 were assigned as differentially expressed. GO enrichment analysis was implemented by the Novogene cloud platform (https://magic.novogene.com) with adjusted p-value < 0.05 and log2 foldchange > 1.
RNA isolation and RT-qPCR
The transcription levels of genes were analysed by RT-qPCR. Cells were collected and immediately frozen in liquid nitrogen. Total RNAs of samples were extracted by Yeast RNAiso Kit (Takara). For cDNAs synthesis, PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) from Takara was utilized with 1 μg of the total RNA as a template based on the manufacturer’s protocol. The reaction was terminated by heating at 85 °C for 5 s. The actin gene ACT1 was selected as an endogenous control for the RT-qPCR to normalize the expression data for each gene. The primers for RT-qPCR used in this study are listed in Table 2. Amplification was performed using 2*SG Fast qPCR Master Mix (High Rox) (Sangon Biotech, China) in the ABI StepOne Plus Real-Time PCR System (ABI, Germany). The thermal cycling conditions for gene amplification were initial denaturation at 95 °C for 30 s, followed by 40 cycles each of denaturation at 95 °C for 3 s and annealing at 60 °C for 30 s. All experiments were independently performed in triplicate. The expression ratio of a gene was analysed using the 2−ΔΔCt method [55]. Statistical analyses of data were performed using Microsoft’s Excel software.
Table 2.
List of genes and primers for RT-qPCR used in this study
| Name | Primer sequence (5ʹ-3ʹ) |
|---|---|
| ACT1 |
F: TATTGAAGTTGAAGCCCTCTGAGC R: CCTTCCGTGTGCAAATGAAACAC |
| CAT |
F: TTCGACAACGCTAATCACGCTAAC R: TCACCTCAAACTCACCGAAAGCT |
| SOD1 |
F: TCGAACAATCCTCCGAAAGCAGC R: CCGTGGGTCTTACCAAATGGGT |
| AHP1 |
F: CTGGAGTCTTAGCTTGTGCTATTCC R: CGTCGATTTTCTCAATGAAAACAGG |
| GLR1 |
F: GGTGCAAAGACCCTTTTGATTGAAG R: GGTCCAGTTAAAGGAAAAATCGCC |
| TRR1 |
F: GGGTATGTTGGCTAACGGTATCG R: GGTCTCGGTAATGATCTCAGTGCC |
| GSH2 |
F: GCGATGTACCCCACCAATTTTGAG R: GCCCAGCCATTCCTGATTTTTGA |
| ZWF1 |
F: AGGGCGACGAGGACAAAGTTC R: GGGCAAAGCTAAGTAGAACAACCTG |
| GND2 |
F: GGTTTCACCGTCGTCGCTTACA R: CGGGATTACCAGCCTTGACCAATA |
| Mxr1 |
F: CCACAGTTGCAATACCAACAATCTC R: CGGAGAAGGAAGCTCTTCTTAGT |
| Prm1 |
F: ACAGGGACAATCTCTGAGTCTGA R: GTTGGAGCACTGTTGGGGTATAT |
| Mit1 |
F: GGTTCAAACTCTGGCAGTTCAGAT R: CCATTGGACATTCCATTGAAACCG |
All ORF sequences can be found from http://PICHIAGENOME.org database
Forward (F) and reverse (R) primer sequences
Acknowledgements
Not applicable.
Abbreviations
- WT
Wild type
- ROS
Reactive oxygen species
- ER
Endoplasmic reticulum
- UPR
The unfolded protein response
- PDI
Protein disulfide isomerase
- GFP
Green fluorescence protein
- AOX1
Alcohol oxidase 1
- GAP
Glyceraldehyde 3-phosphate dehydrogenase gene
- H2O2
Hydrogen peroxide
- GEO
Gene Expression Omnibus
- PCA
Principal component analysis
- GO
Gene Ontology
- BP
Biological process
- CC
Cellular component
- MF
Molecular function
- NOX
NADPH oxidase
Authors’ contributions
NXL, XWY designed the experiments while NXL, RZH performed the experiments in the laboratory. NXL wrote the paper and XWY and YX helped in writing and reviewing the research paper and provided valuable suggestions. All authors read and approved the final manuscript.
Funding
This work is supported by the National Key Research and Development Program of China [Grant number 2021YFC2100203]; the National Natural Science Foundation of China [Grant number 31671799]; the National Natural Science Foundation of China [Grant number 32072162]; the Postgraduate Research & Practice Innovation Program of Jiangsu Province [Grant number KYCX18_1791]; and the National First-Class Discipline Program of Light Industry Technology and Engineering [Grant number LITE2018-09].
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 2.Goncalves AM. Pichia pastoris: a recombinant microfactory for antibodies and human membrane proteins. J Microbiol Biotechnol. 2013;23:587–601. doi: 10.4014/jmb.1210.10063. [DOI] [PubMed] [Google Scholar]
- 3.Cereghino GPL, Cereghino JL, Ilgen C, Cregg JM. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol. 2002;13:329–332. doi: 10.1016/S0958-1669(02)00330-0. [DOI] [PubMed] [Google Scholar]
- 4.Damasceno LM, Pla I, Chang HJ, Cohen L, Ritter G, Old LJ, Batt CA. An optimized fermentation process for high-level production of a single-chain Fv antibody fragment in Pichia pastoris. Protein Expr Purif. 2004;37:18–26. doi: 10.1016/j.pep.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 5.Chung JD. Design of metabolic feed controllers: application to high-density fermentations of Pichia pastoris. Biotechnol Bioeng. 2000;68:298–307. doi: 10.1002/(SICI)1097-0290(20000505)68:3<298::AID-BIT8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 6.Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22:249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- 7.Sarramegna V, Muller I, Mousseau G, Froment C, Monsarrat B, Milon A, Talmont F. Solubilization, purification, and mass spectrometry analysis of the human mu-opioid receptor expressed in Pichia pastoris. Protein Expr Purif. 2005;43:85–93. doi: 10.1016/j.pep.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Yang ZL, Zhang ZS. Codon-optimized expression and characterization of a pH stable fungal xylanase in Pichia pastoris. Process Biochem. 2017;53:80–87. doi: 10.1016/j.procbio.2016.11.022. [DOI] [Google Scholar]
- 9.Norden K, Agemark M, Danielson JA, Alexandersson E, Kjellbom P, Johanson U. Increasing gene dosage greatly enhances recombinant expression of aquaporins in Pichia pastoris. BMC Biotechnol. 2011;11:47. doi: 10.1186/1472-6750-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sha C, Yu XW, Li F, Xu Y. Impact of gene dosage on the production of lipase from Rhizopus chinensis CCTCC M201021 in Pichia pastoris. Appl Biochem Biotechnol. 2013;169:1160–1172. doi: 10.1007/s12010-012-0050-9. [DOI] [PubMed] [Google Scholar]
- 11.Whyteside G, Alcocer MJC, Kumita JR, Dobson CM, Lazarou M, Pleass RJ, Archer DB. Native-state stability determines the extent of degradation relative to secretion of protein variants from Pichia pastoris. PLoS ONE. 2011;6:10. doi: 10.1371/journal.pone.0022692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helian Y, Gai Y, Fang H, Sun Y, Zhang D. A multistrategy approach for improving the expression of E. coliphytase in Pichia pastoris. J Ind Microbiol Biotechnol. 2020;47:1161–1172. doi: 10.1007/s10295-020-02311-6. [DOI] [PubMed] [Google Scholar]
- 13.Tan TW, Lu JK, Nie KL, Deng L, Wang F. Biodiesel production with immobilized lipase: a review. Biotechnol Adv. 2010;28:628–634. doi: 10.1016/j.biotechadv.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Chauhan M, Chauhan RS, Garlapati VK. Evaluation of a new lipase from Staphylococcus sp. for detergent additive capability. Biomed Res Int. 2013;2013:6. doi: 10.1155/2013/374967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seddigi ZS, Malik MS, Ahmed SA, Babalghith AO, Kamal A. Lipases in asymmetric transformations: recent advances in classical kinetic resolution and lipase–metal combinations for dynamic processes. Coord Chem Rev. 2017;348:54–70. doi: 10.1016/j.ccr.2017.08.008. [DOI] [Google Scholar]
- 16.Sha C, Yu XW, Lin NX, Zhang M, Xu Y. Enhancement of lipase r27RCL production in Pichia pastoris by regulating gene dosage and co-expression with chaperone protein disulfide isomerase. Enzyme Microb Technol. 2013;53:438–443. doi: 10.1016/j.enzmictec.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Mo WJ, Wang MZ, Zhan RR, Yu Y, He YG, Lu H. Kluyveromyces marxianus developing ethanol tolerance during adaptive evolution with significant improvements of multiple pathways. Biotechnol Biofuels. 2019;12:15. doi: 10.1186/s13068-019-1393-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh EJ, Wei N, Kwak S, Kim H, Jin YS. Overexpression of RCK1 improves acetic acid tolerance in Saccharomyces cerevisiae. J Biotechnol. 2019;292:1–4. doi: 10.1016/j.jbiotec.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Lin NX, He RZ, Xu Y, Yu XW. Augmented peroxisomal ROS buffering capacity renders oxidative and thermal stress cross-tolerance in yeast. Microb Cell Fact. 2021;20:131. 10.1186/s12934-021-01623-1. [DOI] [PMC free article] [PubMed]
- 20.Zhang P, Zhang WW, Zhou XS, Bai P, Cregg JM, Zhang YX. Catabolite repression of Aox in Pichia pastoris is dependent on hexose transporter PpHxt1 and pexophagy. Appl Environ Microbiol. 2010;76:6108–6118. doi: 10.1128/AEM.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad M, Hirz M, Pichler H, Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol. 2014;98:5301–5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang CB, Ma Y, Miao HB, Tang XH, Xu B, Wu Q, Mu YL, Huang ZX. Transcriptomic analysis of Pichia pastoris (Komagataella phaffii) GS115 during heterologous protein production using a high-cell-density fed-batch cultivation strategy. Front Microbiol. 2020;11:17. doi: 10.3389/fmicb.2020.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. BBA-Mol Cell Res. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russmayer H, Buchetics M, Gruber C, Valli M, Grillitsch K, Modarres G, Guerrasio R, Klavins K, Neubauer S, Drexler H, Steiger M, Troyer C, Al Chalabi A, Krebiehl G, Sonntag D, Zellnig G, Daum G, Graf AB, Altmann F, Koellensperger G, Hann S, Sauer M, Mattanovich D, Gasser B. Systems-level organization of yeast methylotrophic lifestyle. BMC Biol. 2015;13:25. doi: 10.1186/s12915-015-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez S, Navas-Yuste S, Payne AM, Rivera W, Lopez-Estepa M, Brangbour C, Fulla D, Juanhuix J, Fernandez FJ, Vega MC. Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes. Free Radic Biol Med. 2019;141:279–290. doi: 10.1016/j.freeradbiomed.2019.06.025. [DOI] [PubMed] [Google Scholar]
- 26.Martins D, English AM. Catalase activity is stimulated by H2O2 in rich culture medium and is required for H2O2 resistance and adaptation in yeast. Redox Biol. 2014;2:308–313. doi: 10.1016/j.redox.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aksam EB, Jungwirth H, Kohlwein SD, Ring J, Madeo F, Veenhuis M, van der Klei IJ. Absence of the peroxiredoxin Pmp20 causes peroxisomal protein leakage and necrotic cell death. Free Radic Biol Med. 2008;45:1115–1124. doi: 10.1016/j.freeradbiomed.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Souabni H, Machillot P, Baciou L. Contribution of lipid environment to NADPH oxidase activity: influence of sterol. Biochimie. 2014;107:33–42. doi: 10.1016/j.biochi.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Garrigos-Martinez J, Nieto-Taype MA, Gasset-Franch A, Montesinos-Segui JL, Garcia-Ortega X, Valero F. Specific growth rate governs AOX1 gene expression, affecting the production kinetics of Pichia pastoris (Komagataella phaffii) P-AOX1-driven recombinant producer strains with different target gene dosage. Microb Cell Fact. 2019;18:15. doi: 10.1186/s12934-019-1240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang XL, Wang Q, Wang JJ, Bai P, Shi L, Shen W, Zhou M, Zhou XS, Zhang YX, Cai MH. Mit1 transcription factor mediates methanol signaling and regulates the alcohol oxidase 1 (AOX1) promoter in Pichia pastoris. J Biol Chem. 2016;291:6245–6261. doi: 10.1074/jbc.M115.692053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin-Cereghino GP, Godfrey L, de la Cruz BJ, Johnson S, Khuongsathiene S, Tolstorukov I, Yan MD, Lin-Cereghino J, Veenhuis M, Subramani S, Cregg JM. Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol Cell Biol. 2006;26:883–897. doi: 10.1128/MCB.26.3.883-897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takagi S, Tsutsumi N, Terui Y, Kong XY, Yurimoto H, Sakai Y. Engineering the expression system for Komagataella phaffii (Pichia pastoris): an attempt to develop a methanol-free expression system. FEMS Yeast Res. 2019;19:10. doi: 10.1093/femsyr/foz059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pena DA, Gasser B, Zanghellini J, Steiger MG, Mattanovich D. Metabolic engineering of Pichia pastoris. Metab Eng. 2018;50:2–15. doi: 10.1016/j.ymben.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 34.Bodvard K, Peeters K, Roger F, Romanov N, Igbaria A, Welkenhuysen N, Palais G, Reiter W, Toledano MB, Kall M, Molin M. Light-sensing via hydrogen peroxide and a peroxiredoxin. Nat Commun. 2017;8:11. doi: 10.1038/ncomms14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu K, Gao LM, Ul Hassan J, Zhao ZP, Li C, Huo YX, Liu GY. Improving the thermo-tolerance of yeast base on the antioxidant defense system. Chem Eng Sci. 2018;175:335–342. doi: 10.1016/j.ces.2017.10.016. [DOI] [Google Scholar]
- 36.Ask M, Mapelli V, Hock H, Olsson L, Bettiga M. Engineering glutathione biosynthesis of Saccharomyces cerevisiae increases robustness to inhibitors in pretreated lignocellulosic materials. Microb Cell Fact. 2013;12:10. doi: 10.1186/1475-2859-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prouzet-Mauleon V, Monribot-Espagne C, Boucherie H, Lagniel G, Lopez S, Labarre J, Garin J, Lauquin GJM. Identification in Saccharomyces cerevisiae of a new stable variant of alkyl hydroperoxide reductase 1 (Ahp1) induced by oxidative stress. J Biol Chem. 2002;277:4823–4830. doi: 10.1074/jbc.M109614200. [DOI] [PubMed] [Google Scholar]
- 38.MacDiarmid CW, Taggart J, Kerdsomboon K, Kubisiak M, Panascharoen S, Schelble K, Eide DJ. Peroxiredoxin chaperone activity is critical for protein homeostasis in zinc-deficient yeast. J Biol Chem. 2013;288:31313–31327. doi: 10.1074/jbc.M113.512384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant CM, Collinson LP, Roe JH, Dawes IW. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996;21:171–179. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- 40.Zepeda AB, Figueroa CA, Abdalla DSP, Maranhao AQ, Ulloa PH, Pessoa A, Jr, Farias JG. Biomarkers to evaluate the effects of temperature and methanol on recombinant Pichia pastoris. Braz J Microbiol. 2014;45:475–483. doi: 10.1590/S1517-83822014000200014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anelli T, Sannino S, Sitia R. Proteostasis and “redoxtasis” in the secretory pathway: tales of tails from ERp44 and immunoglobulins. Free Radic Biol Med. 2015;83:323–330. doi: 10.1016/j.freeradbiomed.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 42.Qie B, Lyu Z, Lyu L, Liu J, Gao X, Liu Y, Duan W, Zhang N, Du L, Liu K. Sch9 regulates intracellular protein ubiquitination by controlling stress responses. Redox Biol. 2015;5:290–300. doi: 10.1016/j.redox.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mejia-Barajas JA, Montoya-Perez R, Salgado-Garciglia R, Aguilera-Aguirre L, Cortes-Rojo C, Mejia-Zepeda R, Arellano-Plazas M, Saavedra-Molina A. Oxidative stress and antioxidant response in a thermotolerant yeast. Braz J Microbiol. 2017;48:326–332. doi: 10.1016/j.bjm.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuehne A, Emmert H, Soehle J, Winnefeld M, Fischer F, Wenck H, Gallinat S, Terstegen L, Lucius R, Hildebrand J, Zamboni N. Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol Cell. 2015;59:359–371. doi: 10.1016/j.molcel.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 45.Nocon J, Steiger M, Mairinger T, Hohlweg J, Rumayer H, Hann S, Gasser B, Mattanovich D. Increasing pentose phosphate pathway flux enhances recombinant protein production in Pichia pastoris. Appl Microbiol Biotechnol. 2016;100:5955–5963. doi: 10.1007/s00253-016-7363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu K, Qin L, Bai WX, Wang XY, Li F, Ren SC, Gao XP, Chen B, Tong Y, Li J, Li BZ, Yuan YJ, Li C. Multilevel defense system (MDS) relieves multiple stresses for economically boosting ethanol production of industrial Saccharomyces cerevisiae. ACS Energy Lett. 2020;5:572–582. doi: 10.1021/acsenergylett.9b02681. [DOI] [Google Scholar]
- 47.Xu P, Qiao KJ, Stephanopoulos G. Engineering oxidative stress defense pathways to build a robust lipid production platform in Yarrowia lipolytica. Biotechnol Bioeng. 2017;114:1521–1530. doi: 10.1002/bit.26285. [DOI] [PubMed] [Google Scholar]
- 48.Mathur R, Das RP, Ranjan A, Shaha C. Elevated ergosterol protects Leishmania parasites against antimony-generated stress. FASEB J. 2015;29:4201–4213. doi: 10.1096/fj.15-272757. [DOI] [PubMed] [Google Scholar]
- 49.Landolfo S, Zara G, Zara S, Budroni M, Ciani M, Mannazzu I. Oleic acid and ergosterol supplementation mitigates oxidative stress in wine strains of Saccharomyces cerevisiae. Int J Food Microbiol. 2010;141:229–235. doi: 10.1016/j.ijfoodmicro.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 50.Higgins VJ, Beckhouse AG, Oliver AD, Rogers PJ, Dawes IW. Yeast genome-wide expression analysis identifies a strong ergosterol and oxidative stress response during the initial stages of an industrial lager fermentation. Appl Environ Microbiol. 2003;69:4777–4787. doi: 10.1128/AEM.69.8.4777-4787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallstrom BM, Petranovic D, Nielsen J. Altered sterol composition renders yeast thermotolerant. Science. 2014;346:75–78. doi: 10.1126/science.1258137. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez-Vargas S, Sanchez-Garcia A, Martinez-Rivas JM, Prieto JA, Randez-Gil F. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl Environ Microbiol. 2007;73:110–116. doi: 10.1128/AEM.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitichantaropas Y, Boonchird C, Sugiyama M, Kaneko Y, Harashima S, Auesukaree C. Cellular mechanisms contributing to multiple stress tolerance in Saccharomyces cerevisiae strains with potential use in high-temperature ethanol fermentation. AMB Express. 2016;6:107. doi: 10.1186/s13568-016-0285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 55.Jie HF, Qi F, Liu HW, Zou HF, Ahmed MS, Li C. Novel helper factors influencing recombinant protein production in Pichia pastoris based on proteomic analysis under simulated microgravity. Appl Microbiol Biotechnol. 2015;99:653–665. doi: 10.1007/s00253-014-6175-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.