Fig 2. Cytokinesis-blocked micronucleus assay to quantify somatic cell instability frequencies in the trisomic compared to disomic cells from people with mosaicism for a trisomy 21 imbalance.
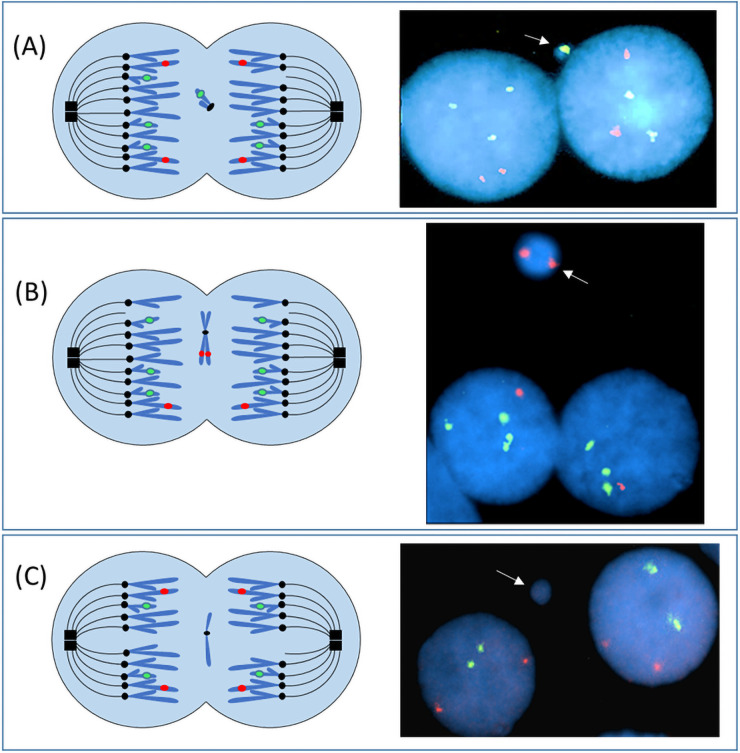
This diagram illustrates one mechanism (chromosome or chromatid lagging) whereby micronuclei can form. (A) During the metaphase of a mitotic division one chromatid from chromosome 21 fails to attach to the spindle fibers. As a result, this chromatid lags behind during the anaphase migration and fails to segregate to the spindle poles (left diagram). Following karyokinesis, the laggard chromosome could be excluded from the daughter cell nuclei and become enclosed in a micronucleus. In the right photomicrograph of a trisomic cell, at least a portion of chromosome 21 was excluded into a micronucleus (white arrow). Only 2 signals for the chromosome 21 probe are present in the right daughter nucleus (loss of a chromosome 21 signal) compared to 3 signals that are present in the left daughter nucleus (RUNX1 probe [21q22; green]; RUNX1T1 probe (8q22; red)]. In panel (B) trisomic binucleates (3 signals for the chromosome 21 probe) are shown (illustration on left; photomicrograph on right) that had loss of one replicated chromosome 8 (both sister chromatids) into a micronucleus, resulting in daughter cells that each had a monosomic imbalance for chromosome 8. (C) A disomic binucleated cell (both primary nuclei have 2 signals for the chromosome 21 and chromosome 8 probes) has a single micronucleus that does not contain chromatin for either the RUNX1 (21q22) or the RUNX1T1 (8q22) loci.