Figure 5.
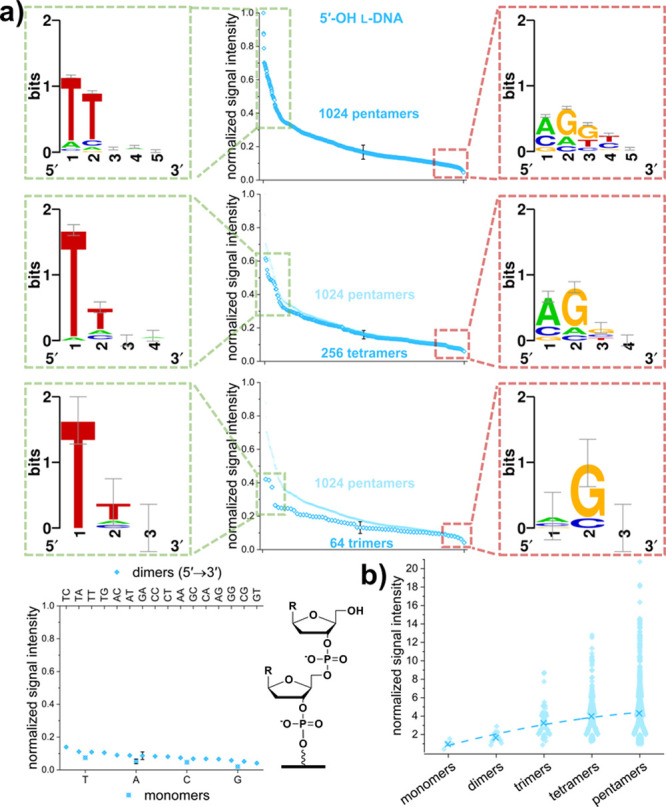
Analysis of 5′-OH l-DNA strand extension data. (a) Fluorescent signal intensities were normalized to the maximum and grouped by length, with representative error bars corresponding to 2× SEM for better visibility. Panels framed in green illustrate sequence patterns for the 10% highest signal intensities, whereas panels framed in red show the data for the 10% lowest. Pentamer data are repeated in subsequent plots for comparison, pointing to the similarity in shape between graphs for differing strand lengths. For monomers and dimers, data are plotted from highest to lowest signal intensity with the corresponding sequence specified on the labeling of the top and bottom x-axis for dimers and monomers, respectively. Next to this plot, the chemical structure of a dimer on the glass surface serves as a guide for identification of differences between the chemical variants tested for initiation. (b) Fluorescent signal intensities normalized to the average of all monomers and clustered according to strand length. Averages for each strand length are indicated by “×”. The dotted line is a second order polynomial fit through the averages.
