Table 2.
Comparison of the proposed method (Method 1), traditional method (Method 2), commercial kit for proteomics (Method 3), and two commercial kits for peptide mapping (Methods 4 and 5).
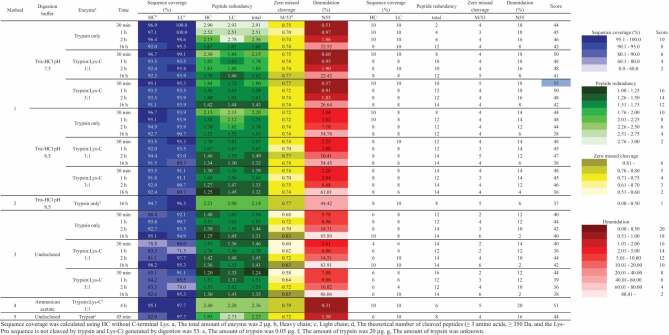 |
Sequence coverage was calculated using HC without C-terminal Lys. a, The total amount of enzyme was 2 µg. b, Heavy chain; c, Light chain; d, The theoretical number of cleaved peptides (≥ 3 amino acids, ≥ 350 Da, and the Lys-Pro sequence is not cleaved by trypsin and Lys-C) generated by digestion was 53. e, The amount of trypsin was 0.05 µg. f, The amount of trypsin was 20 µg. g, The amount of trypsin was unknown.
