Figure 1.
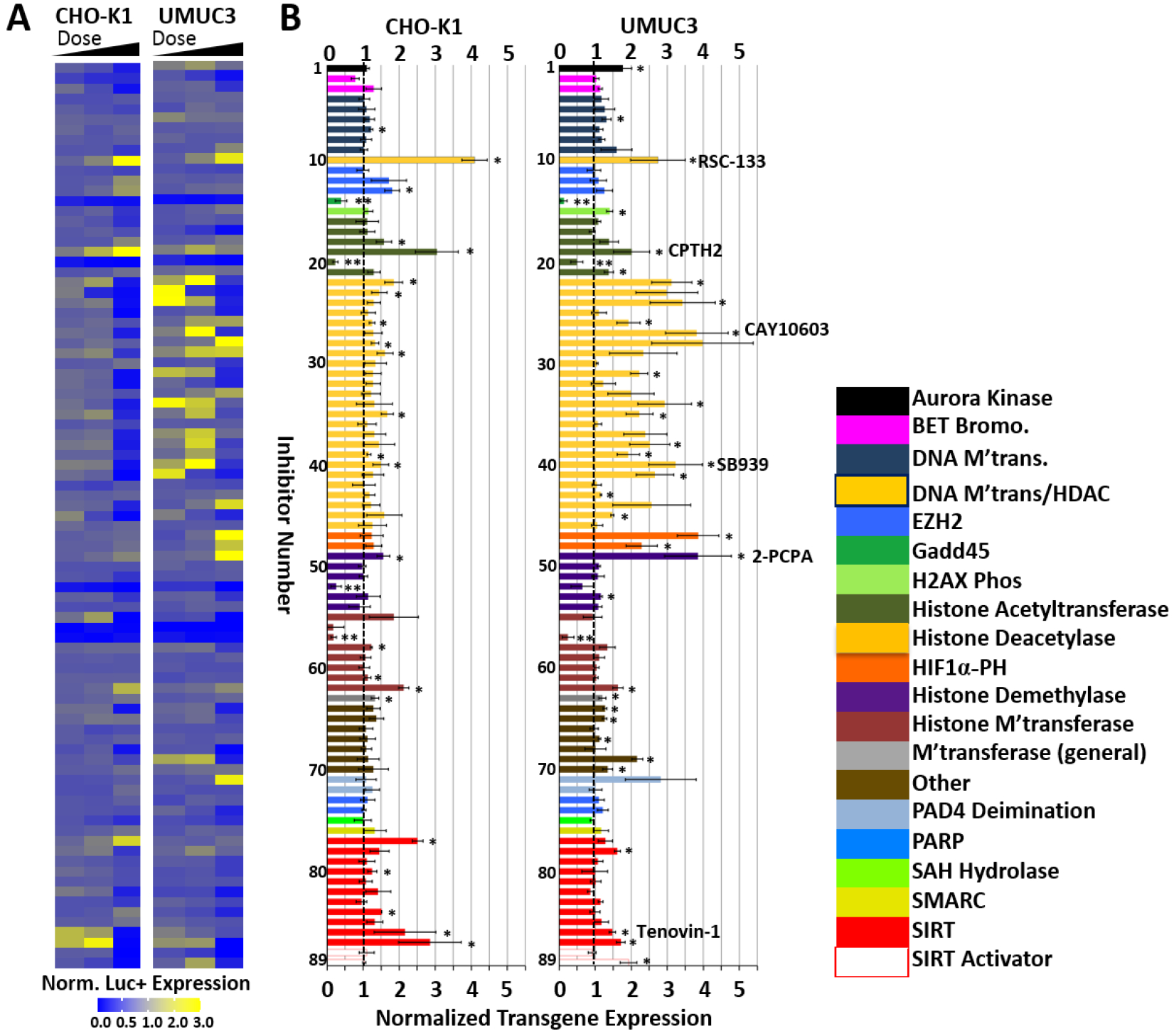
Small-molecule inhibitors of epigenetics modulators were screened to identify leads that enhanced polymer-mediated transgene expression in CHO-K1 cells and UMUC3 bladder cancer cells. A) Heat map indicating the normalized luciferase expression ranges for each of the three doses examined for each inhibitor in the screen. B) Normalized transgene expression for optimal inhibitor dose; color-coding indicates modulator family targeted by the indicated inhibitor. Vertical dashed line indicates point of no enhancement of transgene expression. Inhibitor numbers (1–89) can be referenced in Table S1, supporting information section, for each individual small molecule. Lead inhibitors discussed in more depth are listed. Polyplexes were formed by complexing the N-RDGE polymer with 75 ng EF-Luc plasmid DNA at a 25:1 (wt/wt) ratio. * = Student’s t-test p<0.05 for enhancement in transgene expression relative to DMSO vehicle (no inhibitor) + polyplex control; ** = Student’s T-test p<0.05 for reduction in transgene expression relative to DMSO vehicle (no inhibitor) + polyplex control. Chaetocin (inhibitor #56) was not considered a statistically significant reducer of transgene expression due to its high toxicity at the doses tested.
