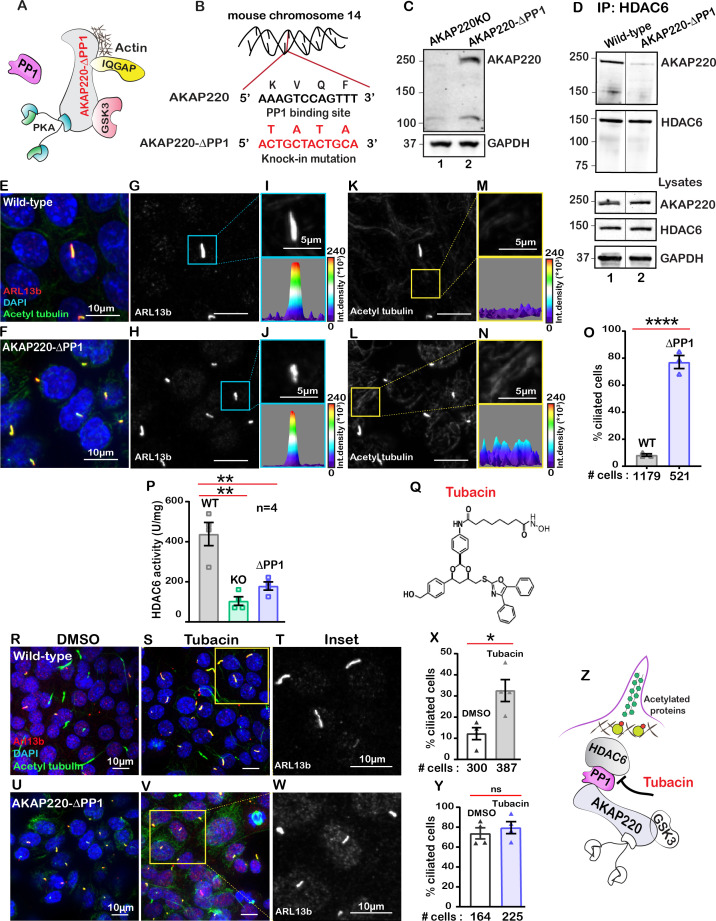
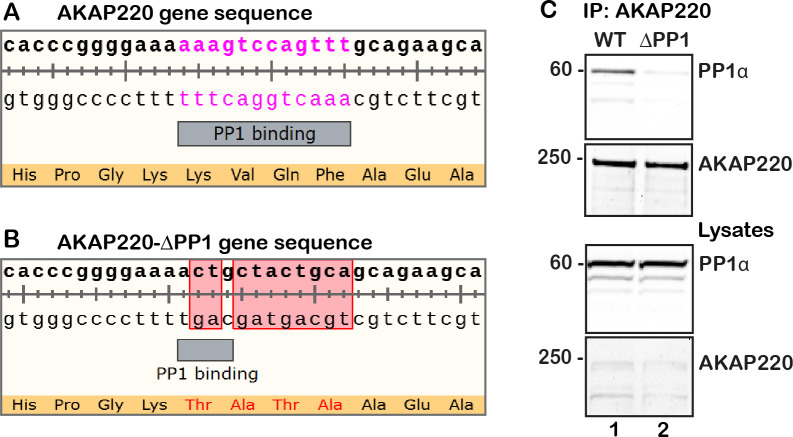
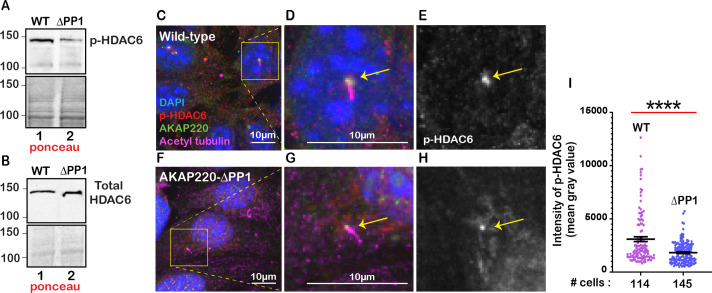
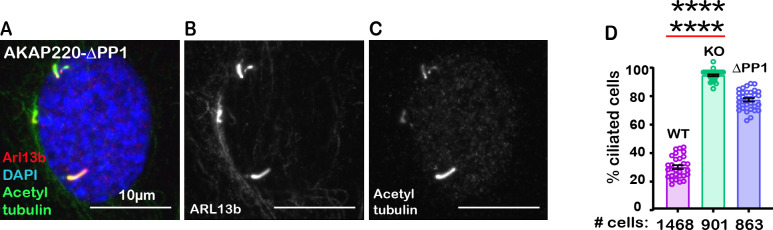
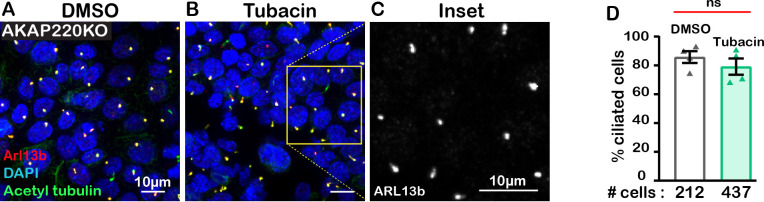
Figure 3. Anchored protein phosphatase one is necessary for HDAC6 activity.
(A) Schematic of AKAP220-ΔPP1. Binding partners are indicated. Gene editing deleted the principal phosphatase binding site (KVQF) on AKAP220. (B) Nucleotide sequencing reveals substitution of the KVxF motif. (C) Immunoblot detection of AKAP220 (top) and GAPDH loading control (bottom) in AKAP220KO (lane 1) and AKAP220-ΔPP1 (lane 2) mIMCD3 cell lysates. (D) Loss of PP1-targeting motif in AKAP220 negatively impacts association with HDAC6. Co-immunoprecipitation studies show that wild-type AKAP220 recruits HDAC6 (lane 1). AKAP220-ΔPP1 recruits less HDAC6 (lane 2). Immunoblot detection of AKAP220 (top), HDAC6 (mid) in cell lysates and GAPDH loading controls (bottom) reveal that equivalent levels of both proteins were present in mIMCD3 cell lysates. (E–O) Immunofluorescent detection of acetyl tubulin (green), Arl13b (red), and DAPI (blue) in (E) wild-type and (F) AKAP220-ΔPP1 cells. Gray scale images of Arl13b in (G) wild-type and (H) AKAP220-ΔPP1 cells. A single enlarged cilium (top) and corresponding three-dimensional surface plots (bottom) from (I) wild-type and (J) AKAP220-ΔPP1 cells. Gray scale images of acetylated tubulin in (K) wild-type and (L) AKAP220-ΔPP1 cells. Enlarged sections from (K and L) (top) and corresponding three-dimensional surface plots (bottom) from (M) wild-type and (N) AKAP220-ΔPP1 cells. (O) Quantification (% ciliated cells) in wild-type (gray) and AKAP220-ΔPP1 (blue). ****p<0.0001, N=3. (P) HDAC6 activity levels (A.U.) in wild-type (gray), AKAP220KO (green) and AKAP220-ΔPP1 (blue) cells as assessed by Bioline’s activity assay. **p<0.01, N=4. (Q) Chemical structure of HDAC6 inhibitor tubacin. (R-Y) Tubacin enhances ciliogenesis in the presence of native AKAP220. Wild-type mIMCD3 cells treated with (R) DMSO or (S) tubacin (2 µM) for 4 hr. Immunofluorescent staining with acetyl tubulin (green), Arl13b (red), and DAPI (blue). (T) Higher magnification gray scale image of Arl13b staining. (X) Quantification (% ciliated cells) in DMSO (white) and tubacin-treated (gray) wild-type cells. *p<0.05, ns=non-significant; N=3. (U–Y) Tubacin has no effect on AKAP220-ΔPP1 cells. (U) DMSO and (V and W) tubacin-treated AKAP220-ΔPP1 cells. (Y) Quantification (% ciliated cells) and analysis as described above in DMSO (white) and tubacin-treated (blue). (Z) Schematic of proposed tubacin mechanism of action on AKAP220-signaling complex. All error bars are s.e.m. p Values were calculated by unpaired two-tailed Student’s t-test. Scale bars (10 µm). Number of cells analyzed indicated below each column.