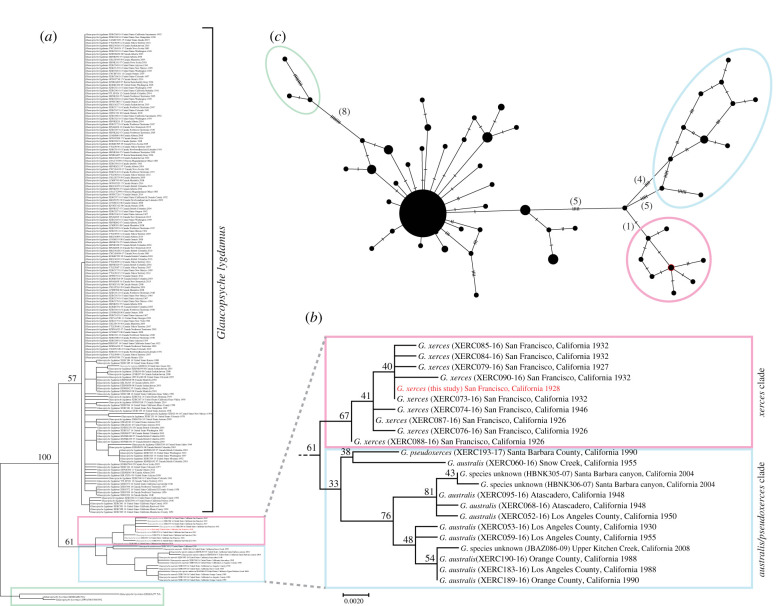
Figure 1.
(a) Phylogenetic tree of Glaucopsyche. The tree was reconstructed by maximum likelihood inference of the CO1 barcoding mitochondrial marker gene. Three G. lycormas sequences were used as an outgroup. Glaucopsyche xerces of this study is highlighted in red. Bootstrap values of main clades are indicated near each node. The detailed tree is presented in electronic supplementary material, figure S3. (b) Expansion of the xerces and australis/pseudoxerces clades of the phylogenetic tree in (a). Bootstrap support values are indicated near each node. Taxon labels indicate species names, BOLD Process IDs, collection locations and collection years. (c) Haplotype network of all Glaucopsyche calculated from the CO1 barcoding marker used in the phylogeny of (a). Mutations are indicated by dashes and numbers in brackets. Ellipses highlight the clusters of G. xerces (pink), G. australis/pseudoxerces (light blue) and G. lycormas (green). Red circle indicates the node of the network that included G. xerces.