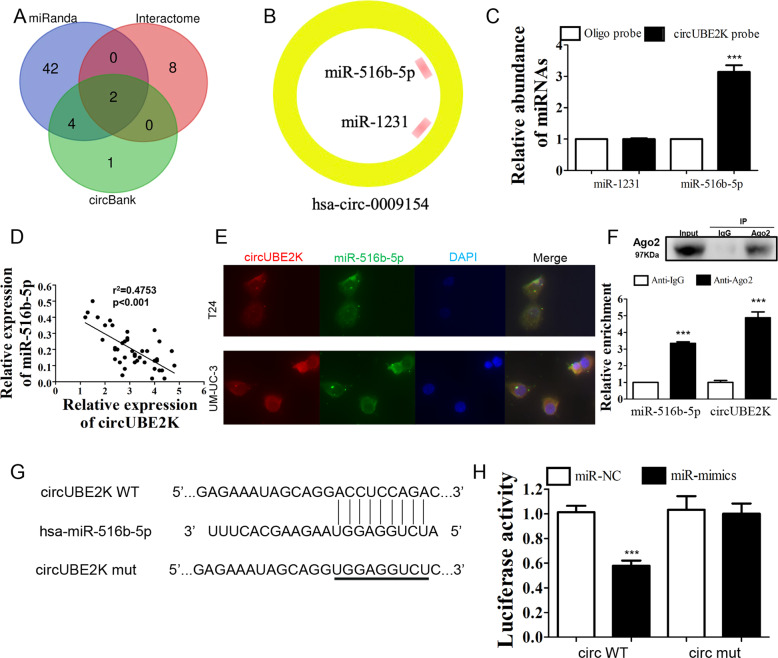
Fig. 3. circUBE2K acted as a sponge for miR-516b-5p in BC cells.
A Venn diagram showing overlapping of potential targets of circUBE2K predicted by miRanda, Circular RNA Interactome and CircBank Database. B Two miRNAs (miR-516b-5p and miR-1231) were predicted by three different prediction software programs. C RNA-pulldown assay followed by qRT-PCR suggested interaction between circUBE2K and miR-516b-5p. Error bars indicate SD (***P < 0.001 vs. other groups). D Scatter plot presented negative correlation between expression level of circUBE2K and miR-516b-5p in BC tissues (n = 42, r2 = 0.4753, P < 0.001). E Co-localization of circUBE2K (Red) and miR-516b-5p (Green). Nuclear staining was performed using DAPI (Blue). F RIP experiments were performed in T24 cells against IgG or Ago2, and the precipitated circUBE2K and miR-516b-5p were detected by qRT-PCR. ***P < 0.001 vs IgG. G Predicted binding site of circUBE2K and miR-516b-5p was demonstrated. The third row showed mutated circUBE2K sequence for further dual-luciferase reporter assay. H Luciferase activity were measured after co-transfection with circUBE2K (circWT)/mutated circUBE2K (circMUT) and miRNA mimics negative control (miR-NC)/miR-516b-5p mimics (miR-mimics) in HEK-293T cells. Error bars indicate SD (***P < 0.001 vs. vehicle).