Introduction
Pancreatic cancer, particularly pancreatic ductal adenocarcinoma (PDAC), is an aggressive disease with a poor prognosis.1 Surgery is the only potential curative treatment, but it is only possible in patients with early-stage disease, and most patients present with non-resectable disease.1 Even in patients who undergo resection, the recurrence rate is very high due to early systemic dissemination.2 Combinatorial chemotherapy remains the standard of care for patients with advanced disease, but responses to it are heterogeneous, and its toxicity limits treatment duration.3–5 Results from clinical trials of single-agent immunotherapy have not proven its efficacy in patients with pancreatic cancer, not even when used with chemotherapy or other immunotherapeutic agents.6–8 A highly immunosuppressive tumour microenvironment (TME) has been postulated as one of the main reasons for the lack of efficacy of immunotherapy for pancreatic cancer.9–11 Therefore, novel strategies that modulate the suppressive TME are urgently needed.
The GI tract is the largest reservoir of microbes, which play important roles in modulating metabolism and immunity through interaction with host cells.12 The discovery of the association between infection with the bacterium Helicobacter pylori and the incidence of gastritis and peptic ulcer disease earned Barry Marshall and Robin Warren the Nobel Prize in Physiology or Medicine in 2005, underscoring the pivotal role of the microbiota in influencing inflammatory conditions which may predispose to cancer.13 The microbial α-diversity is a metric that quantifies the number of different species within a defined sample.14 15 In patients with many malignancies, including colorectal, breast and pancreatic cancer, the α-diversity of the gut microbiota is lower than that in healthy controls.16–18 With respect to clinical outcomes, the gut microbial α-diversity is higher in patients with melanoma that responded to immunotherapy than in those unresponsive.19 In this review, we highlight recent studies analysing microbes present in various compartments (oral cavity, gut, cysts and pancreatic tumours) in patients with pancreatic cancer, as well as murine studies. We also discuss various strategies for microbial modulation. A summary of the published data on enriched microbes at different sites in clinical studies and murine models of pancreatic cancer is included in table 1.
Table 1.
The enriched microbes in the oral cavity, gut and tumours in murine models and human samples
| Host | Study group | Enriched bacteria | Reference |
| Oral microbiota | |||
| Human | Prediagnostic samples, PDAC versus matched controls | Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans | Fan et al 20 |
| Human | PDAC versus healthy controls | Neisseria elongate, Streptococcus mitis | Farrell et al 21 |
| Gut microbiota | |||
| Human | PDAC versus healthy controls | Prevotella, Veillonella, Klebsiella, Selenomonas, Hallella, Enterobacter, Cronobacter | Ren et al 18 |
| Human | PDAC versus healthy controls | Bacteroidetes, Veillonellaceae, Akkermansia, Odoribacter | Half et al 22 |
| Human | PDAC versus healthy controls | Proteobacteria, Synergistetes, Euryarchaeota | Pushalkar et al 23 |
| Mouse | PDAC spontaneous model | Actinobacteria, Deferribacteres, Bifidobacterium pseudolongum | |
| Mouse | PDAC subcutaneous model | Bacteroidetes, Firmicutes | Sethi et al 48 |
| Mouse | PDAC spontaneous model | Bacteroides, Alphaproteobacteria | Mendez et al 26 |
| Tumour microbiota | |||
| Human | PDAC versus healthy controls | Gammaproteobacteria | Geller et al 37 |
| Human | PDAC tumour versus gut | Proteobacteria (Pseudomonas, Elizabethkingia) | Pushalkar et al 23 |
| Human | PDAC | Acinetobacter, Afipia, Corynebacterium, Escherichia, Propionibacterium | Thomas et al 27 |
| Human | PDAC | Gammaproteobacteria, Bacilli, Actinobacteria | Riquelme et al 40 |
| Human | PDAC long-term survivors versus short-term survivors | Pseudoxanthomonas, Streptomyces, Saccharopolyspora, Bacillus clausii | |
| Cyst microbiota | |||
| Human | IPMN, MCN, SCA, pseudocysts | Bacteroides, Escherichia/Shigella, Fusobacterium, Acidaminococcus, Sphingomonas, Bifidobacterium | Li et al 35 |
| Human | Cancerous versus non-cancerous PCNs | Fusobacterium nucleatum, Granulicatella adiacens | Gaiser et al 36 |
IPMN, intraductal papillary mucinous neoplasm; MCN, mucinous cystic neoplasm; PCN, pancreatic cystic neoplasias; PDAC, pancreatic ductal adenocarcinoma; SCA, serous cystadenoma.
Relevance of the oral and gut microbiomes in pancreatic cancer
Several recent studies have analysed the oral and gut microbiomes associated with pancreatic cancer risk and progression.18 20–25 Periodontal disease has been recognised as a risk factor for pancreatic cancer and may be an initiator of oral microbial dysbiosis.25 A prospective population-based nested case–control study demonstrated that the presence of Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans in the oral cavity was indicative of increasing the risk of pancreatic cancer.20 Notably, high levels of plasma antibodies reactive against P. gingivalis corresponded with reduced risk of pancreatic cancer, potentially due to systemic immunity against cancer-associated oral pathogens.24 Another study demonstrated that the oral microbial composition differs in healthy controls and patients with established pancreatic cancer, and the investigators proposed that detection of two microbes in the oral compartment (Neisseria elongata and Streptococcus mitis) can differentiate patients with pancreatic cancer from healthy individuals.21
The gut microbiota associated with pancreatic cancer development has been assessed in a few clinical studies.18 22 A study in China examined a population of patients with PDAC and matched healthy controls.18 Besides a lower α-diversity in the gut microbiome of patients with PDAC versus healthy controls, a unique gut microbial profile was detected in patients with PDAC, including increased abundance of Veillonella, Klebsiella and Selenomonas species and lipopolysaccharide-producing bacteria (Prevotella, Hallella and Enterobacter species) but decreased abundance of Bifidobacterium species and some butyrate-producing bacteria (eg, Coprococcus, Clostridium, Blautia, Flavonifractor and Anaerostipes species). Of note, patients who presented with biliary obstruction had a unique microbiome, suggesting that biliary fluid stasis has a role in the microbial changes found in these patients.18 Another study that compared gut microbes in patients with pancreatic cancer and healthy controls found an increase in Bacteroidetes and a reduction in Firmicutes abundance in patients with pancreatic cancer in two independent cohorts.22 Analysis of the gut microbiotas of patients with PDAC in a third study revealed enrichment in Proteobacteria, Synergistetes and Euryarchaeota species than in matched healthy controls.23
The functional role of the gut microbiome in pancreatic cancer development has been examined in murine studies.23 26 27 Transgenic mice engineered to develop the full spectrum of pancreatic premalignant to malignant lesions exhibited marked temporal changes in their gut microbiota composition as well as bacteria-related metabolites throughout tumourigenesis.23 26 To examine progressive changes in the gut microbiota during pancreatic tumourigenesis, Pushalkar et al 23 longitudinally sampled faeces from the premalignant PDAC mouse model KC (Ptf1aCre, LSL-KrasG12D). Actinobacteria and Bifidobacterium pseudolongum were abundant in KC mice with advanced stages of disease progression.23 Moreover, a clear delay in tumourigenesis was seen in KC mice located in a germ-free environment but not in their littermates raised in a regular mouse housing facility, highlighting the functional importance of the gut microbiota in pancreatic cancer development.23 The mechanisms implicated in tumourigenic induction by the dysbiotic microbiota included activation of Toll-like receptors (TLRs) on immunosuppressive monocytic cells. Antibiotics-based microbial ablation increased CD4+ T-cell polarisation towards a Th1 phenotype and increased cytotoxic CD8+ T cells based on upregulation of T-bet, tumour necrosis factor-α (TNF-α), interferon gamma (IFN-γ), and CD38. Both CD4+ and CD8+ T cells had overexpression of programmed cell death protein 1 (PD-1), and CD44 and CD4+ T cells had overexpression of inducible costimulator (ICOS) and LFA-1 on microbial ablation. They also found that faecal microbial transplants (FMT) from the spontaneous PDAC mouse model KPC (LSL-KrasG12D, LSL-Trp53R172H and Pdx1Cre) into either germ-free mice or antibiotic-treated KC mice significantly accelerated tumour growth in both settings. Bacterial ablation on recipient mice prior to FMT improves colonisation with donor’s stools. This study further showed that infiltration of immunosuppressive myeloid cells and macrophages into the TME decreased with antibiotic-mediated bacterial ablation, which was reversed with murine FMT from mice with PDAC in an orthotopic PDAC murine model.
A second mechanism postulated for the microbial effect in tumourigenesis implicates metabolites, which are well-known host modulators.26 Mendez et al 26 profiled the gut microbial communities in mice throughout tumourigenesis and identified metabolic pathways associated with microbial changes via metagenomics. They used the genetically engineered spontaneous PDAC mouse model KPC to examine microbial and metabolic changes over time.26 They found that the main metabolic pathways enriched in KPC mice were related to the biosynthesis of pyrimidines and polyamines, in particular, those involving putrescine, spermidine and spermine. Polyamine levels were measured in the serum of KPC mice and were found to be increased at 4 months, the time at which most of the KPC mice had advanced pancreatic intraepithelial neoplasia (PanIN).26 To validate this finding, polyamine levels were analysed in serum samples and were found to be higher in the serum of patients with PDAC than in the serum of healthy controls.26 Polyamines, which are mainly produced by intestinal microbiota, are known to induce cellular proliferation by contributing to purine/pyrimidine cellular biosynthesis.28 One of the microbes associated with polyamine metabolism is Lactobacillus. Authors have reported that Lactobacillus rhamnosus is capable of affecting polyamine metabolism and tumour growth in gastric cancer cases.29 The role of bacteria-derived polyamines in pancreatic cancer development has yet to be established. Also, because of their wide repertoire of microbial enzymes, microbes have great potential to influence host metabolism downstream of dietary influences, including metabolism of xenobiotics by altering their toxicity.30
In summary, the oral and gut microbiomes may play an important role in pancreatic cyst biology and cancer initiation and progression through modulation of immune and metabolic pathways. Functional mechanisms implicated in these processes must be further delineated. Nevertheless, sequential analysis of oral and/or gut bacteria, as well as their serum-associated metabolites, could emerge as an inexpensive, non-invasive strategy for early detection of pancreatic cancer. Furthermore, targeting microbial populations associated with increased risk of PDAC may also represent a novel cancer-preventive methodology.
Relevance of the pancreatic intratumoral microbiome
The presence of micro-organisms in tumours has been described multiple times over the past 130 years.31–34 Early pancreatic cystic lesions have been described to possess bacterial DNA.35 36 In examination of different types of pancreatic cystic lesions, such as intraductal papillary mucinous neoplasms (IPMNs), mucinous cystic neoplasms (MCN), serous cystadenomas (SCA) and pseudocysts, most samples had enrichment in Bacteroides spp, Escherichia/Shigella species, Fusobacterium acidaminococcus, Sphingomonas species and Bifidobacterium spp.35 Another study demonstrated that the microbiotas of high-grade IPMNs were commonly found as part of oral microbiome, like F. nucleatum and Granulicatella adiacens; this potential translocation was attributed in part to invasive endoscopic procedures.36
In 2017, Geller et al 37 reported for the first time the presence of bacteria in human PDACs while demonstrating that intratumoral Gammaproteobacteria, among the most common bacteria detected in human pancreatic tumours, reduce the efficacy of chemotherapeutic drugs like gemcitabine. The mechanism for this involves the bacterial production of cytidine deaminase, which is capable of metabolising gemcitabine into its inactive form.37 Other comprehensive studies have revealed the presence of microbiotas in several other tumour types, such as melanoma and breast, lung, ovarian, bone and brain tumours.38 Tumour microbial signatures can also be extracted from publicly available data sets in The Cancer Genome Atlas (TCGA) by filtering out non-human sequences from tumour and blood samples and can be used, through artificial intelligence driven predictive models, to distinguish between cancer and healthy signatures as well as different cancer types.39
Intratumoral bacteria have also been identified in murine PDAC models.23 27 Of note, though, is that Thomas et al 27 reported that genetically engineered mice (KrasG12D/+, PTENlox/+ and Pdx1-Cre) had tumourigenesis delay when raised in a germ-free environment. However, intratumoral microbiotas did not suffer major changes after they were transferred from germ-free to specific pathogen-free housing conditions, suggesting that the local tumour microbial communities in this model may not be as relevant as systemic/gut communities.
More recently, Riquelme et al 40 profiled intratumoral bacteria from patients with resected PDAC and compared short-term and long-term survivors in two geographically distant cohorts. Long-term survivors had greater intratumoral microbial α-diversity than did those who died of the disease within 5 years after resection.40 Overall tumour microbial characterisation revealed a microbial composition similar to the one in human PDAC previously described by Geller et al,37 but unique enrichment in the following microbes was found in tumours from long-term survivors in both independent cohorts studied by Riquelme and colleagues: Pseudoxanthomonas, Streptomyces and Saccharopolyspora and Bacillus clausii. In this study, tumour immunological profiling revealed enhanced immunoactivation of the TME in long-term survivors, which positively correlated with the survivor-enriched microbes, demonstrating a potential role for tumour microbes in altering immune-cell function. Two of the most enriched bacterial species identified in the tumours of long-term survivors have documented immunomodulatory functions. First, Saccharopolyspora spp induce hypersensitivity pneumonitis41 42 and promote a proinflammatory response in lung epithelial cells via activation of protein kinase D1 through the innate immune signal transduction adaptor MyD88.42 Second, B. clausii can mediate immune-cell production of nitrous oxide and IFN-γ, along with increased CD4+ T-cell proliferation in vitro.43 B. clausii spores are very popular probiotics in Europe, branded as Enterogermina.43 In vitro studies have shown that B. clausii can protect enterocytes from rotavirus infection by improving epithelial barrier function and reducing the production of reactive oxygen species and cytokines like interleukin-8 and interferon-β.44 B. clausii can also decreased the toxicity of pathogens like Clostridium difficile and Bacillis cereus through secretion of a serine protease.45 No clinical or preclinical studies have assessed a potential role for B. clausii in the context of cancer.
Another notable microbial population in pancreatic tumours is that of the fungal mycobiome, which was recently reported to be important for pathogenesis.46 Aykut et al 46 showed that the fungal genus Malassezia is abundant in murine and human PDACs and that its depletion with amphotericin B reduced tumour growth in orthotopic and autochthonous pancreatic cancer models. Repopulation of Malassezia but not other genera, such as Candida, Saccharomyces and Aspergillus, promoted tumour growth in orthotopic pancreatic tumour-bearing mice given pretreatment with antifungals.46 Signalling through the mannose-binding lectin (MBL) pathway enabled tumour-associated fungal populations to activate the complement cascade via the C3 molecule.46
Further work is needed to determine the route of colonisation of intestinal bacteria in pancreatic tumours, whether through reflux from the duodenum or via circulation through the bloodstream or lymphatic system.47 Also, it would be important to gain a better understanding of the local conditions that favour a niche for bacteria colonisation in tumours.
Antimicrobials as backbone therapy for pancreatic cancer
Disruption of a dysbiotic microbiota has been associated with beneficial effects and decreased tumour growth in numerous pancreatic cancer murine models.23 27 48 Sethi et al 48 demonstrated that oral treatment with an antibiotic cocktail composed of vancomycin, neomycin, metronidazole, ampicillin and amphotericin B delays the growth of subcutaneous murine PDAC implants by increasing the number of IFN-γ-producing cytotoxic T cells and inhibiting the number of interleukin (IL)-17A and IL-10 producing protumourigenic T cells. Also, Pushalkar et al 23 reported that depletion of the gut microbiota with oral antimicrobials led to decreased tumour size and changes in the immune landscape of the TME in orthotopic PDAC mouse models by eliciting both innate and adaptive responses to reduce myeloid-derived suppressor cells (MDSCs) and increase M1-type macrophages tumour infiltration with suppression of TLR 2/5 signalling while enhancing the number of Th1-type CD4+ and cytotoxic CD8+ T cells. In addition, Thomas et al 27 found that murine gut microbiota ablation with antibiotics dampened tumour growth in genetic PDAC mouse models (KrasG12D/+, PTENlox/+ and Pdx1-Cre). These three independent studies using different PDAC models showed that broad microbial ablation is an effective approach to affect progression of premalignant pancreatic lesions to cancer and PDAC growth.23 27 48 We envision a clinical trial in which chemotherapy is combined with systemic antibiotics for treatment of PDAC. However, known toxic effects associated with long-term use of broad-spectrum antibiotics as well as rise of multidrug resistant bacteria would certainly limit the enthusiasm for such studies.49 50
Elimination of bacteria with targeted local delivery of antibiotics such as ampicillin and chloramphenicol resulted in reduced tumour growth in a subcutaneous colon carcinoma mouse model (MC26) as shown by Geller et al.37 Local intratumoral release of antibiotics was achieved through the use of an implantable microdevice with a standard biopsy needle that can release microdoses of single agents or combinations of therapeutic drugs.51 Increased levels of apoptosis as measured using cleaved caspase 3 staining were observed in tumours only when the implanted microdevice released a combination of gemcitabine and antibiotics, thus highlighting the specific role of intratumoral bacteria in opposing chemotherapy activity.37 Analogous to this, in a colon carcinoma xenograft mouse model, abrogation of the protumourigenic Fusobacterium load via treatment with metronidazole slowed tumour growth as well.52 Localised delivery of antibiotics using local release devices would be less toxic and more effective for systemic use, potentially representing a promising local therapeutic strategy to be combined with systemic antitumoral therapies.37 53
Despite the promising data on antimicrobial use for PDAC described previously, antibiotics have been associated with worse outcomes for other cancer types. Antibiotic use prior to immune checkpoint inhibitors treatment has been associated with worse survival in melanoma, lung and other cancers.54–56 Meta-analyses of published clinical data have also shown that antibiotic usage in patients with cancer (melanoma, lung, renal and head and neck carcinomas) receiving immune checkpoint inhibitors leads to poorer survival.57 Several preclinical studies have also reported on the negative effects of concomitant use of antibiotics and immunotherapy. Iida et al 58 reported a detrimental effect of depletion of the commensal microbiota in a subcutaneous colorectal cancer mouse model (MC38), as treatment with an antibiotic regimen consisting of vancomycin, imipenem and neomycin resulted in decreased efficacy of CpG oligonucleotide immunotherapy and platinum-based chemotherapy. Similarly, in a subcutaneous sarcoma mouse model (MCA-205), Vétizou et al 59 observed decreased potency of inhibition of the activity of the immune checkpoint molecule cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) on ablation of the commensal microbiota with the use of antibiotics such as ampicillin, colistin and streptomycin. Using the same antibiotic cocktail, Routy et al 60 observed reduced efficacy of blocking another immune checkpoint target, PD-1 blockade, in reducing tumour growth in subcutaneous mouse models of sarcoma (MCA-205) and melanoma (RET) when combined with antibiotics.
From the information in the previous two paragraphs, antibiotics may have different impacts depending on the tumour type, the concomitant therapies and the presence or absence of specific bacteria in the cancer cells as in the example of patients with colorectal cancer, in whom antibiotics only play an antitumoral effect when tumour cells contain Fusobacterium.52 Therefore, identifying the microbial profiles present across tumours in different patient populations is important to elucidate their differential effects. If antibiotic treatment in patients with cancers like sarcomas and melanoma results in increased tumour growth, microbial populations in these settings may be beneficial for the host and may be needed for therapeutic responses of tumours.58–60 In patients with pancreatic cancer, however, because most studies have reported antitumoral effect on antimicrobial treatment, microbial enrichment associated with this cancer type may contribute to its pathogenesis, explaining why its elimination may play a beneficial role.23 27 48 Future studies must evaluate factors shaping the overall effect of the tumour microbiota, including variables like diversity, location, the microenvironment and host factors.
Additionally, certain microbes can generate cross-reactive T cells owing to molecular mimicry between tumour and microbial antigens which may trigger antitumor immunity through antigen presentation by major histocompatibility complex class I (MHC-I).61 However, a recent study showed that pancreatic cancer cells have downregulation of MHC-I molecules on their surfaces for evasion of immunosurveillance through upregulation of autophagy, and inhibition of MHC I improves therapeutic outcomes by synergising with immune checkpoint blockade.62 This highlights the importance of ascertaining the balance between eliminating harmful microbiotas and enriching beneficial microbiotas, which can improve antitumor immunity.
Another emerging strategy for depleting pathogenic intratumoral bacteria is the use of a special class of highly selective viruses known as lytic bacteriophages or phages, which can selectively infect and lyse certain bacteria and have been tested for their capacity to shift the gut microbiota in murine models.53 63 Phage display libraries have been shown to target specific organs in vivo, and they can produce bystander cytotoxicity in tumour cells.64–66 In preliminary studies, researchers explored the utility of phages for delivery of anticancer drugs to pancreatic cancers and demonstrated early viability of successful targeting of pancreatic tumour cells.67–69 Whereas substantial data demonstrate the effect of phages on regulation of microbial population dynamics, evidence of their direct effects on human health is limited. Technical limitations in therapeutically using phages, such as dosing, route of administration, tolerance by the host immune system, and specificity to pancreatic cancer microbes and tumour antigens, remain to be addressed before this strategy can be widely adopted.
Fecal microbial transplants as backbone therapy for pancreatic cancer
The potential of the whole gut microbiota to modulate the pancreatic tumour microbiota and outcomes has been explored by several groups using preclinical models.23 40 46 48
To approximate the human scenario, Riquelme et al 40 performed human faecal microbial transplantation (hFMT) in mice that later received orthotopic tumour cell implantation to generate a PDAC humanised microbial mouse model. This study demonstrated that about a quarter of the human PDAC microbial composition but not the adjacent normal tissue overlapped with the human donors' gut microbiota, suggesting the existence of a microbial cross-talk between the gut and pancreatic tumours.40 The same study also demonstrated that the human donors’ gut bacteria were efficiently transferred to the gut in these mice.40 Moreover, the tumour microbial composition in humanised microbial mice was also differentially modulated by transplants from different donors.40 In accordance with previous murine studies, hFMT using samples obtained from healthy controls resulted in slower tumour growth and a more modest reversal of TME immunosuppression than did hFMT with samples from short-term survivors, whereas the most potent reversal of TME immunosuppression and tumour growth was induced by hFMT using samples from long-term survivors of PDAC with no evidence of disease.40 Of note, this effect was lost when antibiotics were given after the transplant or when cytotoxic T cells were depleted.40 This demonstrated the ability of hFMT to positively affect PDAC tumours by modulating the gut/tumour microbial axis along with the immune system. All of this evidence demonstrates that FMT can modulate the gut and tumour microbiotas, the immune activation status and outcomes, paving the way for FMT to be used in combination with other treatment modalities, such as chemotherapy and immunotherapy, for pancreatic cancer and perhaps other tumour types.
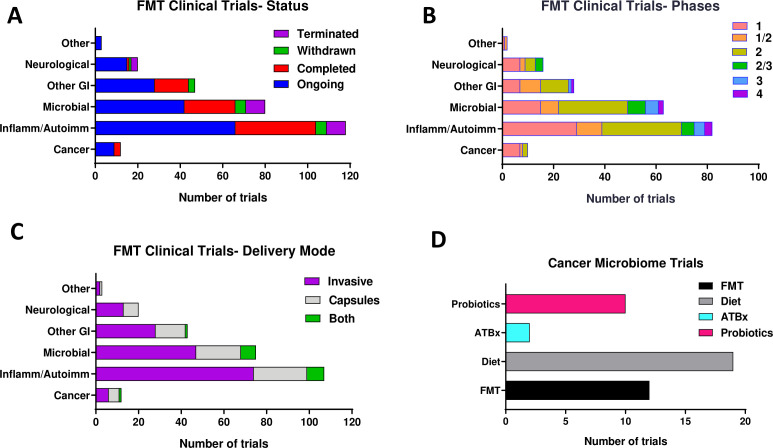
FMT is effectively used in clinical practice for treatment of conditions like recurrent C. difficile infection, and several ongoing clinical trials at different phases of development are using FMT for treatment of autoimmune, inflammatory and metabolic conditions (figure 1A–C and online supplemental table S1).70 71 The use of FMT as a treatment option for cancer is gradually attracting more attention. Several early-phase clinical trials of hFMT are currently testing its role in cancer treatment responses and to reduce treatment-associated toxic effects (figure 1D and online supplemental table S1). Ongoing and future clinical studies will be required to establish the safety and efficacy of FMT in modulating the human pancreatic tumour microbiome and enhancing the immune response to ultimately improve survival.
Figure 1.
Overview of clinical FMT trials by status (A), clinical phase (B), delivery mode (C) and cancer (D) reported in ClinicalTrials.gov as of September 2020. autoimm, autoimmune; FMT, faecal microbial transplant; inflamm, inflammatory.
gutjnl-2019-319807supp001.xlsx (20.8KB, xlsx)
Single or consortia bacteriotherapy
Bacteriotherapy, consisting of oral administration of either a single or a consortium of bacterial species, is a potential strategy for more targeted manipulation of the microbiome than FMT. Early studies in animals looked at the toxicity of spores derived from a non-pathogenic strain of C. novyi, a bacterium that can germinate in necrotic and hypoxic regions of tumours.72 73 Other studies evaluated the roles of specific gut bacterial species in augmenting the efficacy of immunotherapy for tumours.59 74 Bacteroides spp, particularly Bacteroides fragilis and B. thetaiotaomicron, identified in the gut microbiota in a fibrosarcoma mouse model, are capable of enhancing CTLA-4-based immunotherapy.59 Bifidobacterium spp, identified in the gut microbiota of a melanoma murine model, induced antitumourigenic immune responses alone and, more potently, in combination with another immune checkpoint molecule, programmed cell death-ligand 1 (PD-L1).74 Engineered non-pathogenic bacteria also have been studied for targeted delivery of therapeutic agents like antibodies and chemopreventive metabolites to determine their ability to be enriched in tumours.75–77 Nonetheless, no bacterial species have emerged as being therapeutic for pancreatic cancer, and bacteriotherapy for this cancer remains largely unexplored. However, being mindful of the scenarios in which addition of a single bacterium may result in decreased gut diversity, which is usually associated with poor cancer outcomes, is important. Thus, supplementation of single-species bacteriotherapy with FMT may be needed to provide the advantages of both methodologies.
Conclusions
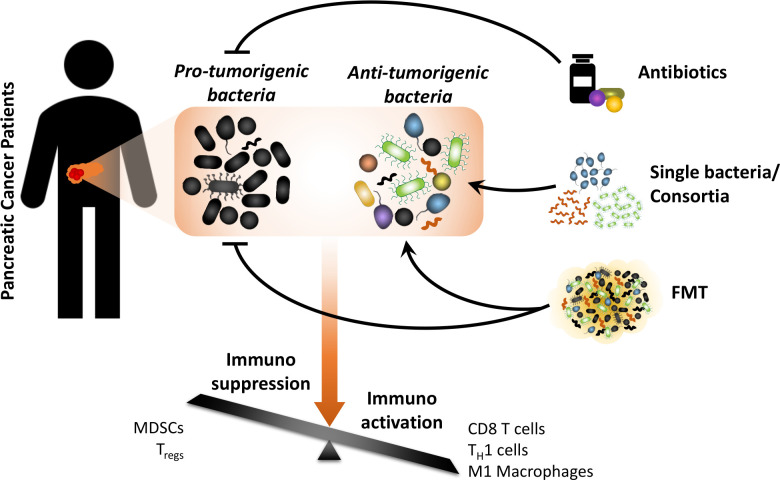
Studies that aim to target the PDAC-associated gut and tumour microbiotas may employ three strategies: (1) elimination of protumourigenic bacteria from the host through the use of antibiotics, (2) enhancement of immunoactivation by delivery of single or multiple microbial species and (3) ‘normalisation’ of the dysbiotic gut and tumour environment in patients with PDAC using whole FMT (figure 2). Ultimately, all of these strategies have the common goal of shifting the balance from an immunosuppressive TME to an immunoactivated one. Furthermore, identification and better understanding of the mechanisms employed by downstream effector molecules (eg, host or microbial metabolites) mediating microbial responses as biomarkers and potentially as therapeutic targets may also be very important. In summary, modulation of the microbiome may emerge as a supplement for existing cancer therapies with the main goal of increasing their efficacy by reversing immunosuppression. Because most functional studies reported thus far have been conducted using animal models, we hope to get more information from upcoming clinical trials targeting the microbiome.
Figure 2.
Diagram showing different approaches to microbiota modulation for cancer treatment. The overall goal is to shift the tumour microenvironment from an immunosuppressive to an immune-activated state. FMT, faecal microbial transplant; MDSC, myeloid-derived suppressor cell; TH1, type I helper; Treg, regulatory T cell.
Footnotes
Contributors: FM designed, wrote and reviewed the article. VC wrote and reviewed the article.
Funding: VC is supported by NCI T32 in Cancer Biology, MD Anderson Cancer Centre. FM is supported by NCI 1R37CA237384-01A1, CPRIT RP200173, V Foundation Translational Award, Andrew Sabin Family Fellowship and Philantropic Moonshot Programmes (MDACC). We also acknowledge the MD Anderson Research Medical Library.
Competing interests: None declared.
Provenance and peer review: Commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not required.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2. Rhim AD, Mirek ET, Aiello NM, et al. Emt and dissemination precede pancreatic tumor formation. Cell 2012;148:349–61. 10.1016/j.cell.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Conroy T, Desseigne F, Ychou M, et al. Folfirinox versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011;364:1817–25. 10.1056/NEJMoa1011923 [DOI] [PubMed] [Google Scholar]
- 4. Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 2013;369:1691–703. 10.1056/NEJMoa1304369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neoptolemos JP, Stocken DD, Bassi C, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA 2010;304:1073–81. 10.1001/jama.2010.1275 [DOI] [PubMed] [Google Scholar]
- 6. O'Reilly EM. Durvalumab with or without tremelimumab for patients with metastatic pancreatic ductal adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol 2019;5:1431–8. 10.1001/jamaoncol.2019.1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kamath SD, Kalyan A, Kircher S, et al. Ipilimumab and gemcitabine for advanced pancreatic cancer: a phase Ib study. Oncologist 2020;25:e808–15. 10.1634/theoncologist.2019-0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Royal RE, Levy C, Turner K, et al. Phase 2 trial of single agent ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother 2010;33:828–33. 10.1097/CJI.0b013e3181eec14c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feig C, Gopinathan A, Neesse A, et al. The pancreas cancer microenvironment. Clin Cancer Res 2012;18:4266–76. 10.1158/1078-0432.CCR-11-3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. von Bernstorff W, Voss M, Freichel S, et al. Systemic and local immunosuppression in pancreatic cancer patients. Clin Cancer Res 2001;7:925s–32. [PubMed] [Google Scholar]
- 11. Liyanage UK, Moore TT, Joo H-G, et al. Prevalence of regulatory T cells is increased in peripheral blood and tumor microenvironment of patients with pancreas or breast adenocarcinoma. J Immunol 2002;169:2756–61. 10.4049/jimmunol.169.5.2756 [DOI] [PubMed] [Google Scholar]
- 12. Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med 2019;216:20–40. 10.1084/jem.20180448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mégraud F. A humble bacterium sweeps this year's Nobel Prize. Cell 2005;123:975–6. 10.1016/j.cell.2005.11.032 [DOI] [PubMed] [Google Scholar]
- 14. Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120–3. 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goedert JJ, Jones G, Hua X, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J Natl Cancer Inst 2015;107. 10.1093/jnci/djv147. [Epub ahead of print: 01 Jun 2015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 2013;105:1907–11. 10.1093/jnci/djt300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ren Z, Jiang J, Xie H, et al. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget 2017;8:95176–91. 10.18632/oncotarget.18820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan X, Alekseyenko AV, Wu J, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2018;67:120–7. 10.1136/gutjnl-2016-312580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farrell JJ, Zhang L, Zhou H, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012;61:582–8. 10.1136/gutjnl-2011-300784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Half E, Keren N, Reshef L, et al. Fecal microbiome signatures of pancreatic cancer patients. Sci Rep 2019;9:16801. 10.1038/s41598-019-53041-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pushalkar S, Hundeyin M, Daley D, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov 2018;8:403–16. 10.1158/2159-8290.CD-17-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Michaud DS, Izard J, Wilhelm-Benartzi CS, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 2013;62:1764–70. 10.1136/gutjnl-2012-303006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Michaud DS, Joshipura K, Giovannucci E, et al. A prospective study of periodontal disease and pancreatic cancer in US male health professionals. J Natl Cancer Inst 2007;99:171–5. 10.1093/jnci/djk021 [DOI] [PubMed] [Google Scholar]
- 26. Mendez R, Kesh K, Arora N, et al. Microbial dysbiosis and polyamine metabolism as predictive markers for early detection of pancreatic cancer. Carcinogenesis 2020;41:561–70. 10.1093/carcin/bgz116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomas RM, Gharaibeh RZ, Gauthier J, et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis 2018;39:1068–78. 10.1093/carcin/bgy073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tofalo R, Cocchi S, Suzzi G. Polyamines and gut microbiota. Front Nutr 2019;6:16. 10.3389/fnut.2019.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Russo F, Orlando A, Linsalata M, et al. Effects of Lactobacillus rhamnosus GG on the cell growth and polyamine metabolism in HGC-27 human gastric cancer cells. Nutr Cancer 2007;59:106–14. 10.1080/01635580701365084 [DOI] [PubMed] [Google Scholar]
- 30. Koppel N, Maini Rekdal V, Balskus EP. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017;356. 10.1126/science.aag2770. [Epub ahead of print: 23 Jun 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Russell W. An address on a characteristic organism of cancer. Br Med J 1890;2:1356–60. 10.1136/bmj.2.1563.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rous P. A sarcoma of the fowl transmissible by an agent separable from the tumor cells. J Exp Med 1911;13:397–411. 10.1084/jem.13.4.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wuerthelecaspe V, Alexanderjackson E, Anderson JA, et al. Cultural properties and pathogenicity of certain microorganisms obtained from various proliferative and neoplastic diseases. Am J Med Sci 1950;220:638–48. 10.1097/00000441-195022060-00006 [DOI] [PubMed] [Google Scholar]
- 34. Aries V, Crowther JS, Drasar BS, et al. Bacteria and the aetiology of cancer of the large bowel. Gut 1969;10:334–5. 10.1136/gut.10.5.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li S, Fuhler GM, Bn N, et al. Pancreatic cyst fluid harbors a unique microbiome. Microbiome 2017;5:147. 10.1186/s40168-017-0363-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gaiser RA, Halimi A, Alkharaan H, et al. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut 2019;68:2186–94. 10.1136/gutjnl-2018-317458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Geller LT, Barzily-Rokni M, Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017;357:1156–60. 10.1126/science.aah5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nejman D, Livyatan I, Fuks G, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020;368:973–80. 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poore GD, Kopylova E, Zhu Q, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020;579:567–74. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Riquelme E, Zhang Y, Zhang L, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 2019;178:795–806. 10.1016/j.cell.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marx JJ, Emanuel DA, Dovenbarger WV, et al. Farmer's lung disease among farmers with precipitating antibodies to the thermophilic actinomycetes: a clinical and immunologic study. J Allergy Clin Immunol 1978;62:185–9. 10.1016/0091-6749(78)90105-7 [DOI] [PubMed] [Google Scholar]
- 42. Kim Y-I, Park J-E, Brand DD, et al. Protein kinase D1 is essential for the proinflammatory response induced by hypersensitivity pneumonitis-causing thermophilic actinomycetes Saccharopolyspora rectivirgula. J Immunol 2010;184:3145–56. 10.4049/jimmunol.0903718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Urdaci MC, Bressollier P, Pinchuk I. Bacillus clausii probiotic strains: antimicrobial and immunomodulatory activities. J Clin Gastroenterol 2004;38:S86–90. 10.1097/01.mcg.0000128925.06662.69 [DOI] [PubMed] [Google Scholar]
- 44. Paparo L, Tripodi L, Bruno C, et al. Protective action of Bacillus clausii probiotic strains in an in vitro model of rotavirus infection. Sci Rep 2020;10:12636. 10.1038/s41598-020-69533-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ripert G, Racedo SM, Elie A-M, et al. Secreted compounds of the probiotic Bacillus clausii strain O/C inhibit the cytotoxic effects induced by Clostridium difficile and Bacillus cereus toxins. Antimicrob Agents Chemother 2016;60:3445–54. 10.1128/AAC.02815-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aykut B, Pushalkar S, Chen R, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019;574:264–7. 10.1038/s41586-019-1608-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomas RM, Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol 2020;17:53–64. 10.1038/s41575-019-0242-7 [DOI] [PubMed] [Google Scholar]
- 48. Sethi V, Kurtom S, Tarique M, et al. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology 2018;155:33–7. 10.1053/j.gastro.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barbosa TM, Levy SB. The impact of antibiotic use on resistance development and persistence. Drug Resist Updat 2000;3:303–11. 10.1054/drup.2000.0167 [DOI] [PubMed] [Google Scholar]
- 50. Bonomo RA. Multiple antibiotic-resistant bacteria in long-term-care facilities: an emerging problem in the practice of infectious diseases. Clin Infect Dis 2000;31:1414–22. 10.1086/317489 [DOI] [PubMed] [Google Scholar]
- 51. Jonas O, Landry HM, Fuller JE, et al. An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Sci Transl Med 2015;7:284ra57. 10.1126/scitranslmed.3010564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017;358:1443–8. 10.1126/science.aal5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Duerkop BA, Clements CV, Rollins D, et al. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci U S A 2012;109:17621–6. 10.1073/pnas.1206136109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Elkrief A, El Raichani L, Richard C, et al. Antibiotics are associated with decreased progression-free survival of advanced melanoma patients treated with immune checkpoint inhibitors. Oncoimmunology 2019;8:e1568812. 10.1080/2162402X.2019.1568812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huemer F, Rinnerthaler G, Westphal T, et al. Impact of antibiotic treatment on immune-checkpoint blockade efficacy in advanced non-squamous non-small cell lung cancer. Oncotarget 2018;9:16512–20. 10.18632/oncotarget.24751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pinato DJ, Howlett S, Ottaviani D, et al. Association of prior antibiotic treatment with survival and response to immune checkpoint inhibitor therapy in patients with cancer. JAMA Oncol 2019;5:1774–8. 10.1001/jamaoncol.2019.2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang X-Z, Gao P, Song Y-X, et al. Antibiotic use and the efficacy of immune checkpoint inhibitors in cancer patients: a pooled analysis of 2740 cancer patients. Oncoimmunology 2019;8:e1665973. 10.1080/2162402X.2019.1665973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342:967–70. 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079–84. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–7. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 61. Fluckiger A, Daillère R, Sassi M, et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science 2020;369:936–42. 10.1126/science.aax0701 [DOI] [PubMed] [Google Scholar]
- 62. Yamamoto K, Venida A, Yano J, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020;581:100–5. 10.1038/s41586-020-2229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hsu BB, Gibson TE, Yeliseyev V, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe 2019;25:803–14. 10.1016/j.chom.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Landon LA, Deutscher SL. Combinatorial discovery of tumor targeting peptides using phage display. J Cell Biochem 2003;90:509–17. 10.1002/jcb.10634 [DOI] [PubMed] [Google Scholar]
- 65. Trepel M, Stoneham CA, Eleftherohorinou H, et al. A heterotypic bystander effect for tumor cell killing after adeno-associated virus/phage-mediated, vascular-targeted suicide gene transfer. Mol Cancer Ther 2009;8:2383–91. 10.1158/1535-7163.MCT-09-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature 1996;380:364–6. 10.1038/380364a0 [DOI] [PubMed] [Google Scholar]
- 67. Asar MC, Franco A, Soendergaard M. Phage display selection, identification, and characterization of novel pancreatic cancer targeting peptides. Biomolecules 2020;10. 10.3390/biom10050714. [Epub ahead of print: 05 May 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bedi D, Gillespie JW, Petrenko VA. Selection of pancreatic cancer cell-binding landscape phages and their use in development of anticancer nanomedicines. Protein Eng Des Sel 2014;27:235–43. 10.1093/protein/gzu020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang T, Narayanaswamy R, Ren H, et al. Phage-derived protein-mediated targeted chemotherapy of pancreatic cancer. J Drug Target 2018;26:505–15. 10.1080/1061186X.2017.1405424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bakken JS, Borody T, Brandt LJ, et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 2011;9:1044–9. 10.1016/j.cgh.2011.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012;143:913–6. 10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- 72. Diaz LA, Cheong I, Foss CA, et al. Pharmacologic and toxicologic evaluation of C. novyi-NT spores. Toxicol Sci 2005;88:562–75. 10.1093/toxsci/kfi316 [DOI] [PubMed] [Google Scholar]
- 73. Krick EL, Sorenmo KU, Rankin SC, et al. Evaluation of Clostridium novyi-NT spores in dogs with naturally occurring tumors. Am J Vet Res 2012;73:112–8. 10.2460/ajvr.73.1.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084–9. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chowdhury S, Castro S, Coker C, et al. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med 2019;25:1057–63. 10.1038/s41591-019-0498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer 2010;10:785–94. 10.1038/nrc2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ho CL, Tan HQ, Chua KJ, et al. Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nat Biomed Eng 2018;2:27–37. 10.1038/s41551-017-0181-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-319807supp001.xlsx (20.8KB, xlsx)