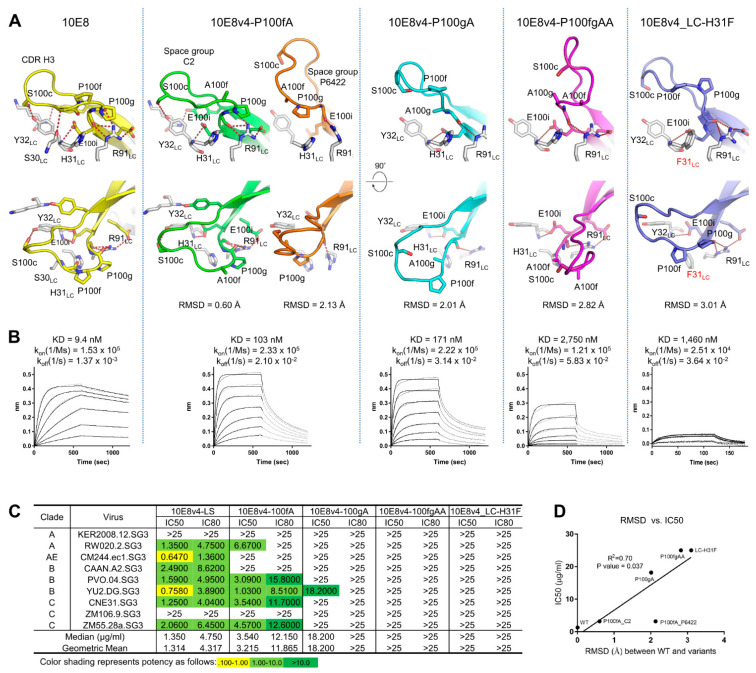
Figure 5.
Active conformation similarity of 10E8 variants involving CDR H3 di-proline motif mutants or motif-interacting residues correlates with neutralization potency. (A) Different conformations of the CDR H3s were shown with RMSDs of Cα between the variants and 10E8 structure. Hydrogen bonds between the residues in the CDR H3 and the light chains were shown in dotted lines. (B) Binding kinetics of Fab molecules of 10E8v4 variants to T117-F MPER scaffold measured by BLI. (C) Neutralization potency and breadth of 10E8v4 variants against select viruses. (D) Correlation between RMSD and median IC50.