Figure 4.
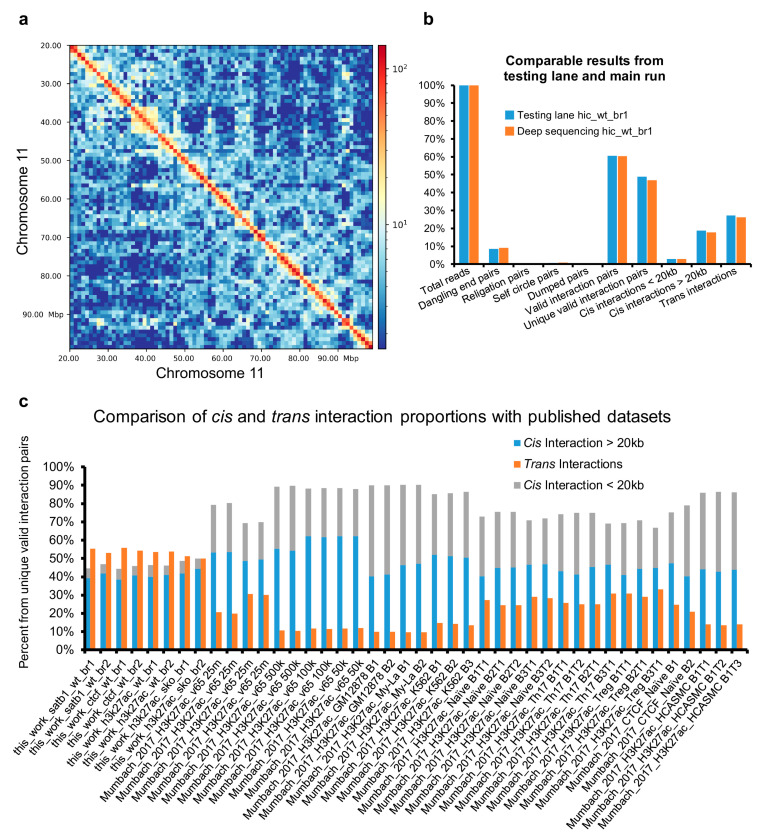
Overview of HiChIP and Hi-C data output. (a) Low-scale sequencing runs can aid in accurately estimating the quality of the prepared DNA libraries. Here, the Hi-C heatmap of murine thymocytes revealed the high-order chromatin organization even at a very low sequencing depth. The sample visualized had only 2.5 million total sequencing reads. (b) The low-scale sequencing runs highly correlated with the final deep sequencing. (c) The optimized protocol yielded a very low portion of short-range (<20 kbp) interactions while providing similar numbers of long-range cis and trans interactions. The comparison to HiChIP libraries produced by the group developing the original HiChIP protocol is provided.