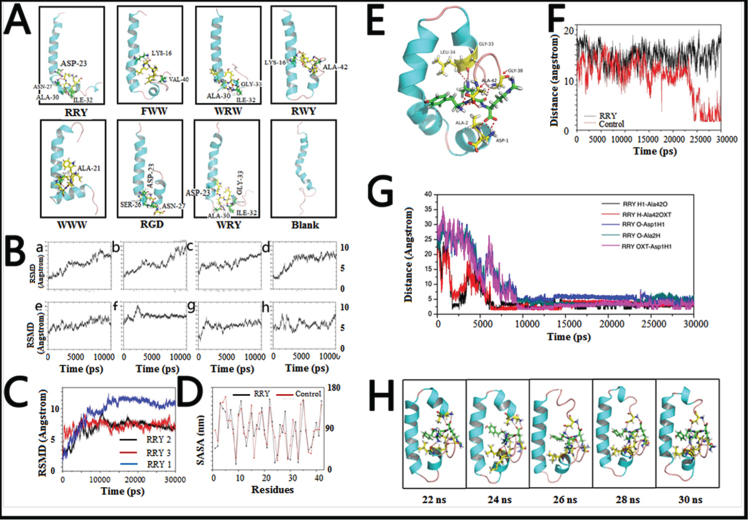
Fig. 2.
Small peptides bind to Aβ1–42 to form a stable structure in silico. A) The final conformations of the small peptides-Aβ1–42 complexes of MD simulation were relatively constant compared to the dramatically changed conformations of Aβ1–42 monomer. B) Root-mean-square deviations (RMSDs) were calculated to investigate the stability of the small peptide- Aβ1–42 complexes. Most complexes remained stable at the last 3 ns of the whole 10 ns simulation. C) RMSDs of the Aβ1–42-RRY complex. The RMSD of Aβ1–42 remained steady at approximately 7 Å or 7 Å in three repeated simulations, indicating that the complex is relatively stable. The red, blue, and black were three repeating trajectories for the MD simulations. D) The SASA of each residue of Aβ1–42 in the presence (black) and absence (red) of RRY. The greater values revealed the weaker hydrophobicity of the residues. E) The conformations of the RRY- Aβ1–42 complexes of MD simulation were relatively constant. It showed that the residues Asp1, Ala2, Gly33, Leu34, Gly38, and Ala42 in Aβ1–42 form hydrogen bonds with RRY. According to the stability of hydrogen bonds, we further emphatically selected five bonds formed by the three Aβ residues above: Asp-1, Ala-2, and Ala-42, to conduct time dependence analysis, the details of which were illustrated in Fig. 2G. F) The distance between Asp23 and Lys28 of Aβ1–42. The distance between Asp23 and Lys28 remained constant at approximately 15 nm when RRY is bound to Aβ1–42 (black), which suggested the absence of the Asp23-Lys28 salt bridge. The distance between Asp23 and Lys28 shows a sharp decrease in Aβ1–42 without RRY bound (red), which suggested the formation of the Asp23-Lys28 salt bridge. G) The key hydrogen bond distances between Aβ1–42 and RRY with time dependence. The five lines in the graph respectively representative the distance between: (i) the hydrogen atom (H) at the amino terminal of RRY and the double bond oxygen atom (O) of the Ala-42 carboxyl group of Aβ (black); (ii) the hydrogen atom (H) on RRY’s first carbon atom and single bond oxygen atom (OXT) of Aβ’s Ala-42 carboxyl group (red); (iii) the hydrogen bond formed by the oxygen atom (O) of RRY and the amino terminal hydrogen atom (H) of Asp-1 of Aβ (blue); (iv) the oxygen atom (O) of RRY and the amino terminal hydrogen atom H of Ala-2 of Aβ (green); (v) RRY’s single-bond oxygen atom O with the hydrogen atom (H) at the amino terminal of Asp-1 of Aβ (purple). The distance between the residues in Aβ1–42 and RRY stays under 3.5 Å and remains stable, which indicated the formation of strong hydrogen bonds. H) The conformation of Aβ1–42 during the MD simulation in the presence RRY. The conformation of Aβ1–42 was stable with RRY bound Aβ1–42.