Table 2.
Parameter settings for simulation
| (A) For structures inferred from three K562 single cells | ||||||||||
| Type | GSM # |

|
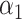
|
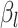
|
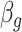
|

|
Sequencing depth | # True 0 positions | Range of 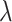 values values |
|
| K562 T1 | 2109974 | 5.6 | −1 | 0.9 | 0.9 | 0.9 | 6800 | 91 | 0.90–16.07 | |
| K562 T2 | 2109985 | 6.3 | −1 | 0.9 | 0.9 | 0.9 | 12,000 | 91 | 0.89–34.41 | |
| K562 T3 | 2109993 | 6.7 | −1 | 0.9 | 0.9 | 0.9 | 13,410 | 91 | 0.87–50.31 | |
| (B) For a structure generated from the helix model | ||||||||||
| Type |

|
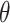
|

|
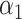
|
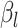
|
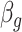
|
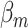
|
Sequencing depth | # true 0 positions | Range of 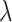 values values |
| Helix T1 | 0.04 |
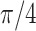
|
1.0 | −1 | 0.1 | 0.1 | 0.1 | 6637 | 91 | 0.83–44.84 |
| Helix T2 | 0.04 |
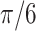
|
1.1 | −1 | 0.1 | 0.1 | 0.1 | 6702 | 91 | 0.91–49.80 |
| Helix T3 | 0.04 |
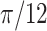
|
1.2 | −1 | 0.1 | 0.1 | 0.1 | 6437 | 91 | 1.02–53.96 |
