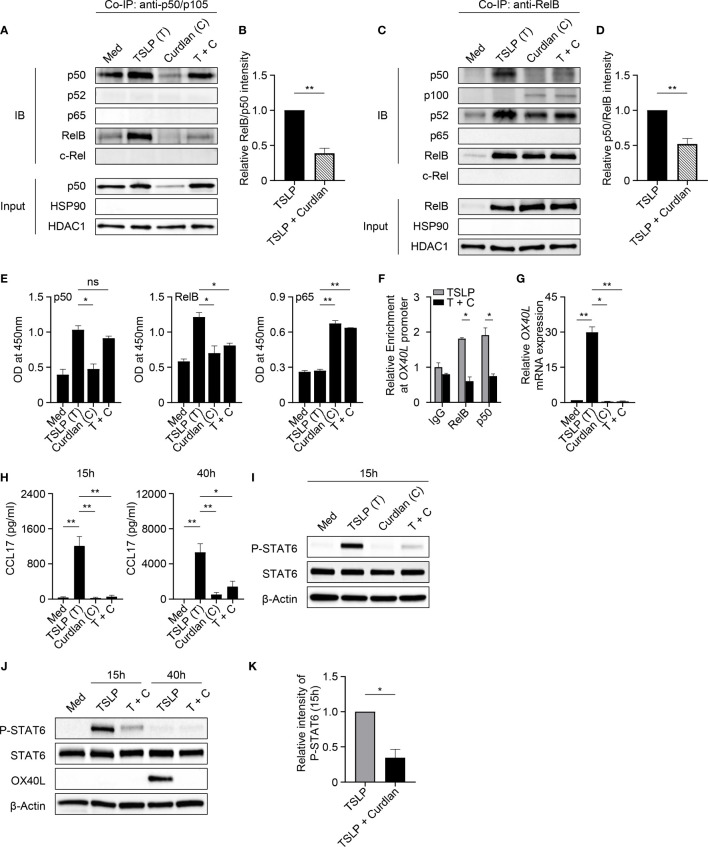
Figure 4.
Dectin-1 Stimulation Inhibits TSLP-Induced OX40L and CLL17 Expression. (A–D) Nuclear proteins from mDCs treated with Med, TSLP and/or curdlan for 40h were used for Co-IP assay. Anti-p50/p105 (A, B) or anti-RelB (C, D) was used as pull-down antibody. Proteins in the immunoprecipitants were analyzed by immunoblotting (IB). In input controls of nuclear proteins, HDAC1 was probed as loading control and HSP90 was probed to investigate the possible cytoplasmic contamination. Representative Western blot data (A, C) and summarized data (B, D) from three independent experiments are shown. The intensity of co-immunoprecipitated protein RelB (A, B) or p50 (C, D) was first normalized to that of IP protein p50 (A, B) or RelB (C, D) in immunoprecipitants, and then relative intensity in TSLP + Curdlan (T + C) group was obtained by comparison with TSLP group. (E) DNA binding of NF-κB subunits in nuclear extracts of mDCs from different donors (n = 3) treated with indicated reagents was measured by ELISA. (F) ChIP assay demonstrates the relative enrichment of RelB and p50 at the region around the κB-like sequences of OX40L promoter in response to TSLP or TSLP and curdlan (T + C). Summarized data from three independent experiments are shown. (G) Quantitative PCR for OX40L mRNA in mDCs from different donors (n = 3) after incubation with indicated reagents for 40h. OX40L expression is normalized to GAPDH and set at 1 in mDCs cultured in Med. (H) CCL17 in culture supernatants of mDCs (n = 4) after incubation with indicated reagents for 15h or 40h was measured by ELISA. (I–K) Whole cell lysates of mDCs incubated with indicated reagents for 15h or 40h were used for immunoblotting. Representative Western blot data (I, J) and summarized data (K) from different donors (n = 3) after 15h stimulation are shown. The intensity of P-STAT6 was first normalized to that of STAT6 in the same group, and then relative intensity was obtained by comparison with Med group. Data in (B, D–H, K) are represented as mean ± SD. Significance was determined using paired t test (two-tailed) for (B, D, K) and one-way ANOVA with Tukey multiple comparisons test for (E, G, H), and two-way ANOVA with Tukey multiple comparisons test for (F). *p < 0.05, **p < 0.01 for the comparison between groups. ns, not significant.