Figure 3 .
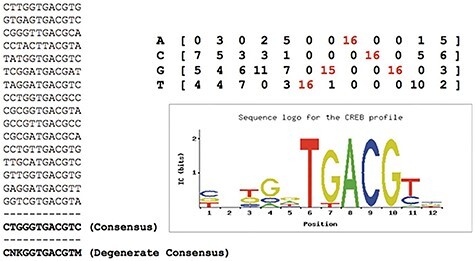
A set of binding sites recognized by the same TF (CREB) [65]. It shows how multiple sequences recognize the same transcription factor (CREB). First, Zambelli built their ‘consensus’ (bottom left) by counting the frequency of each nucleic acid in the sequence [65]. And ‘consensus’ (bottom left) with the highest frequency of nucleotides at each position to indicate the motifs they form a ‘degenerate’ consensus, which includes nucleotides that have no obvious preference position (K = G or T; M = A or C; N = any nucleotide; according to IUPAC codes [105]). Besides, motifs can be converted into an alignment matrix of the nucleotide frequency (top right) by dividing each column by the number of sites used, as well as a ‘sequence logo’ (bottom left) [106] showing nucleotide conservation and corresponding information.
