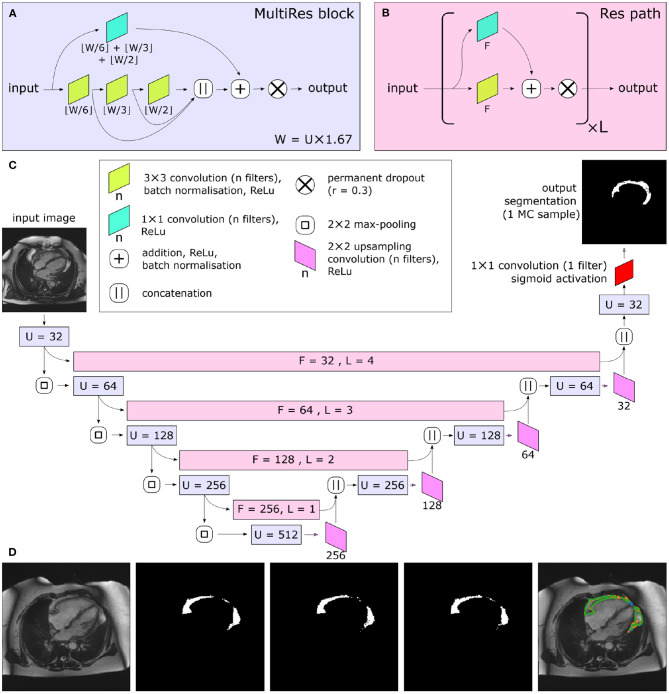
Figure 2.
Central illustration. Summary of model architecture used in the present study. The MultiRes blocks (A) form the encoder and decoder arms of the network. The number of filters used throughout the different components of the block is parameterized by U. The encoder and decoder arms are joined by Res paths (B), which are parameterized by F and L. They are formed of L repeating units, and their convolutional components each have F filters. The complete network is shown in (C). In (C), the colors indicate the placement of MultiRes blocks (A) and Res paths (B), while the hyperparameters used in each instance of the blocks are indicated as overlaid text. Because of the permanently active dropout components, each prediction the network makes is equivalent to a Monte Carlo (MC) sample. (D) shows three such samples drawn based on the same input image. Note the disagreement at the edges of the segmented regions, particularly clear as shown on the overlay (far right). Images reproduced with permission of UK Biobank.