Abstract
Standardized protocols for wastewater-based surveillance (WBS) for the RNA of SARS-CoV-2, the virus responsible for the current COVID-19 pandemic, are being developed and refined worldwide for early detection of disease outbreaks. We report here on lessons learned from establishing a WBS program for SARS-CoV-2 integrated with a human surveillance program for COVID-19. We have established WBS at three campuses of a university, including student residential dormitories and a hospital that treats COVID-19 patients. Lessons learned from this WBS program address the variability of water quality, new detection technologies, the range of detectable viral loads in wastewater, and the predictive value of integrating environmental and human surveillance data. Data from our WBS program indicated that water quality was statistically different between sewer sampling sites, with more variability observed in wastewater coming from individual buildings compared to clusters of buildings. A new detection technology was developed based upon the use of a novel polymerase called V2G. Detectable levels of SARS-CoV-2 in wastewater varied from 102 to 106 genomic copies (gc) per liter of raw wastewater (L). Integration of environmental and human surveillance data indicate that WBS detection of 100 gc/L of SARS-CoV-2 RNA in wastewater was associated with a positivity rate of 4% as detected by human surveillance in the wastewater catchment area, though confidence intervals were wide (β ~ 8.99 ∗ ln(100); 95% CI = 0.90–17.08; p < 0.05). Our data also suggest that early detection of COVID-19 surges based on correlations between viral load in wastewater and human disease incidence could benefit by increasing the wastewater sample collection frequency from weekly to daily. Coupling simpler and faster detection technology with more frequent sampling has the potential to improve the predictive potential of using WBS of SARS-CoV-2 for early detection of the onset of COVID-19.
Keywords: Wastewater based surveillance, SARS-CoV-2, Wastewater, Concentration, Detection, Sampling
Graphical abstract
1. Introduction
The current COVID-19 pandemic has inspired novel methods for monitoring impending outbreaks and for tracking the spread of the disease among human populations. Although COVID-19 is spread primarily through direct person-to-person contact and airborne routes (through inhalation), studies have shown that the RNA of the virus, SARS-CoV-2, is found in feces and in sanitary sewage (i.e., wastewater) (Holshue et al., 2020; Nghiem et al., 2020; Pan et al., 2020; Wu et al., 2020; Xiao et al., 2020; Zhang et al., 2020), with shedding occurring from some pre-symptomatic and asymptomatic individuals at levels of 102 to 107 genomic copies (gc)/mL in feces and 102 to 105 gc/mL in urine (Jones et al., 2020). This is significant, as measurement of SARS-CoV-2 RNA in wastewater can serve as a method of early detection, leading to enhanced warning of an impending outbreak (Ahmed et al., 2020a; La Rosa et al., 2020a; La Rosa et al., 2020b; Lamers et al., 2020; Mao et al., 2020), usually about a week (Medema et al., 2020a; Peccia et al., 2020) or two (Randazzo et al., 2020) before human surveillance using clinical diagnoses can detect infection among the population. As a result, measurement of infectious agents through wastewater-based surveillance (WBS) can empower decision-makers by providing early warnings of outbreaks (Betancourt et al., 2021; Brouwer et al., 2018; Lesimple et al., 2020; Mallapaty, 2020; Shulman et al., 2014; Trottier et al., 2020), which can be a key factor in guiding critical efforts to contain the spread of communicable infectious diseases such as COVID-19.
In addition to serving as an early detection system for disease transmission, advantages of WBS include accounting for individuals who are underestimated by clinical surveillance (Bivins et al., 2020; Kumar et al., 2020; Tang et al., 2020; Wölfel et al., 2020; Ye et al., 2016) because asymptomatic individuals and/or those with mild symptoms may not seek clinical screening and formal diagnoses of the disease (Lodder and de Roda Husman, 2020; Mallapaty, 2020). WBS is a noninvasive method since it identifies buildings and not individuals. This is especially relevant because outbreaks like the COVID-19 pandemic can provoke social stigma and discriminatory behavior toward individuals who may have contracted the virus, which may cause some to conceal the illness and avoid testing to prevent stigmatization (Bhanot et al., 2021). Wastewater viral concentrations represent collective viral shedding by the community at a point in time, providing a temporal distribution of SARS-CoV-2 infection that requires fewer measurements than human surveillance systems. Thus, compared to human surveillance, which requires intense testing of individual humans, WBS is a relatively cost-effective method that requires fewer measurements to assess contributions from a community. Due to these advantages, WBS for SARS-CoV-2 has been implemented around the world as part of wastewater based epidemiologic (WBE) efforts that are aimed at using wastewater detection methods to track infection among communities (Ahmed et al., 2020b; Daughton, 2020; Hamouda et al., 2021; Haramoto et al., 2020; Medema et al., 2020b; Polo et al., 2020; Sims and Kasprzyk-Hordern, 2020).
Although there are many advantages of WBS, its use and implementation need optimization. This includes establishing relationships between WBS measures and the incidence of human illness, improvements in detection sensitivity and quantification, and better strategies for sample collection, concentration, and detection (Kitajima et al., 2020). In this study we implemented a WBS program to evaluate the RNA of SARS-CoV-2 in wastewater. The objective here is to report on lessons learned from establishing a wastewater monitoring program that is integrated with human surveillance of COVID-19 and has the goal of informing decisions needed for timely containment of disease transmission. One important deviation of WBS from traditional wastewater systems design is that measurements are taken much closer to the origin of the source of contamination. Traditionally sewage systems are designed for the conveyance of wastewater, to transport sewage from the buildings to the wastewater plant. The wastewater treatment plant is where the sewage is treated to improve its quality before discharge and thus the quality of wastewater typically focuses on evaluating the wastewater characteristics at the treatment plant. Outside of forensic-based illicit chemical tracking studies (Bannwarth et al., 2019; Centazzo et al., 2019), there are limited data that evaluate water quality characteristics of the sewage upstream from the wastewater plant. One new aspect of the WBS SARS-CoV-2 virus monitoring is that it has focused efforts on evaluating water quality upstream, within the building scale and within the cluster scale (which collects sewage from a group of buildings). Thus, we aim to describe lessons learned from: measuring wastewater quality characteristics closer to the point of initial discharge into the sewage system, concentrating samples for viruses, and detecting SARS-CoV-2 within the concentrates. The unique features of this work include the development of a new qPCR detection strategy for wastewater and the comparisons between SARS-CoV-2 levels in wastewater and corresponding clinical cases.
2. Methods
A WBS monitoring program was established for a university with well-defined municipal water supply characteristics and an elaborate human surveillance monitoring program. The WBS monitoring program was implemented during the Fall 2020 academic semester (August to December). The WBS monitoring program was separated into three components: wastewater sampling, sample processing (e.g., concentration), and SARS-CoV-2 detection. Details of the study site and of the methods employed for each of these components are described below.
2.1. Study site
The WBS study site was the University of Miami (UM), located in Miami-Dade County, Florida, USA. Tap water was the source of wastewater to the sampling sites as the site is characterized by dedicated sewers with no storm water inflows. The source of the tap water was groundwater treated using an enhanced lime softening process which increases the pH of the water (yearly average pH of treated water of 9.1) and decreases the alkalinity (and thus the buffering capacity) (average alkalinity of 51 mg/L as CaCO3). In addition, the tap water was chlorinated using chloramines (combination of chlorine gas and ammonia) with the goal of maintaining a residual at the point of use of at least 0.2 mg/L. To confirm the interconnections of the sewer lines, building and sewer construction drawings were evaluated and sewer lines were dye tested.
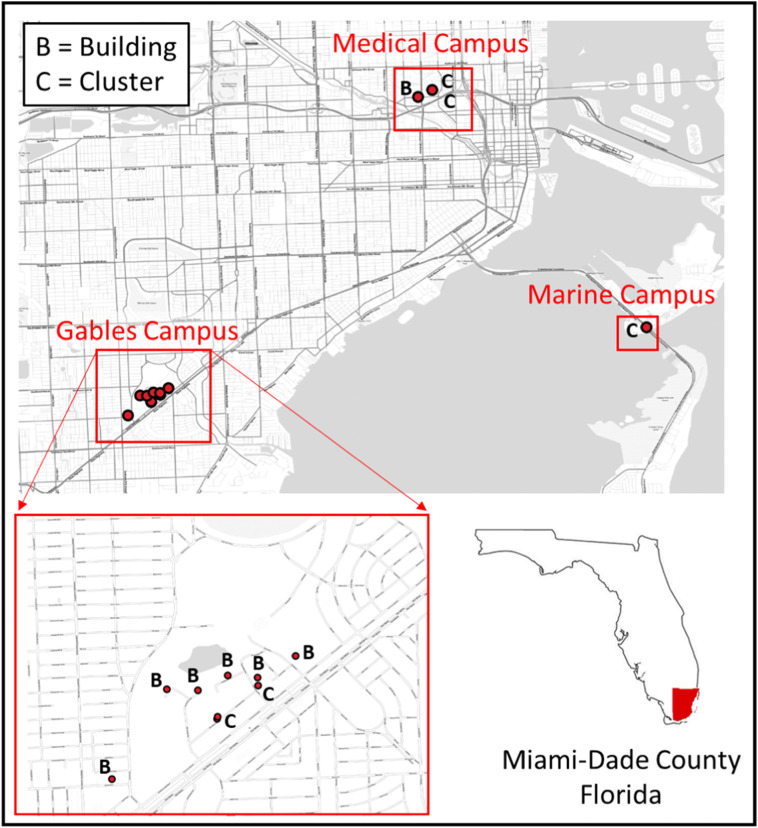
UM is a private research university with a population of more than 34,000 students, faculty, and staff. The three largest campuses include: (1) the Coral Gables campus, which includes most of the teaching and dormitory facilities for the undergraduate student population; (2) the Rosenstiel School of Marine and Atmospheric Science (RSMAS) campus, which predominantly houses research and administrative buildings that focus on studies of marine systems; and (3) the Miller School of Medicine (MSoM) campus, which houses research and administrative facilities plus the University's medical school, the Sylvester Comprehensive Cancer Center (SCCC), and University-owned clinics and hospital. The hospital (UM UHealth Tower, UMHT) treats COVID-19 patients. Samples were collected from manholes or lift stations that serviced a total of 5 clusters of buildings (C1 through C5) on each of the university campuses and from a total of 7 individual buildings (B1 through B7) at the Gables and Miller School campuses (Fig. 1 ). Occupancies and numbers of buildings contributing to each sampling site are provided in Table S-1 located in the supplemental text.
Fig. 1.
Location of building and cluster sampling sites at each of the university campuses.
2.2. Human surveillance program
UM has made tremendous efforts in adapting and responding to the unprecedented events of COVID-19. The University of Miami Health System (UHealth) rapidly built a robust SARS-CoV-2 testing program (Nimer et al., 2020); the University also implemented a human surveillance program through a Testing, Tracing and Tracking (3-T) program. This human surveillance initiative aims to monitor changes in infection burden that may require changes in public health strategy for the UM community.
During the Fall 2020 semester, this human surveillance program included screening of UM students who chose to live on campus and/or attend classes in person. Students who lived on campus were screened once every 10 days and students who lived off-campus and visited campus were screened once every 14 days. These requirements were initiated consistently for student residents effective August 16 and for non-resident students on September 10, 2021. A total of 55,186 tests were conducted on students during the Fall 2020 semester (August 16, 2020 through January 3, 2021). All testing for COVID-19 was conducted via mid-nasal swab followed by PCR-based diagnosis. Each week, a total of 3800 students were screened. These data for resident students were disaggregated by residence hall. Using these data daily positive rate (# of cases per 1000 tested subjects) was computed for each residential hall. Data were interpreted to identify “hot spots” in the community and to establish potential mitigation measures (isolation and/or quarantine) for affected groups of individuals. This human surveillance program was submitted to the University's Internal Review Board (IRB), where it was deemed not to be human subjects' research based on Office for Human Research Protections guidelines. Data reporting was based upon numbers of cases per building with no specific identification of individuals, and hence was not subjected to IRB. Additionally, the names of buildings were de-identified for purposes of reporting results from the study outside of the University.
Human surveillance samples collected through the UM 3-T program were processed at the UM Mailman Center. The Mailman Center, located on the MSoM campus, houses the core microbiology laboratory as well as several research laboratories. Clinical samples sent to the Mailman Center were placed in viral transport media or universal transport media. They were processed using a Perkin Elmer (PE) assay (Perkin Elmer, 2021a; Perkin Elmer, 2021b) which included nucleic acid extraction via the PE Chemagic. A Thermo Fisher Applied Biosystems QuantStudio 7 or Applied Biosystems 7500 Fast was used for RT-qPCR analyses of clinical samples.
2.3. Wastewater sampling
Wastewater was collected at two sewage scales, one corresponding to individual buildings and the other corresponding to clusters of buildings. Individual buildings were serviced by manholes, while clusters of buildings were typically serviced by lift stations. There were no wastewater treatment plants on campus. Wastewater from campus is received by the city-wide sewage collection network which is then treated with effluent discharged to either deep well or ocean outfall. Regular sampling occurred weekly from the campus sewage collection network, on Wednesday mornings from 7:30 am to 10:30 am, starting on September 30, 2020, and continuing through the summer of 2021. The data in this report covers regular sampling through December 16, 2020 (a total of 12 sampling days), with 6 to 12 samples collected each sampling day. Of note, human surveillance data corresponding to this time period extended through January 6, 2021. Early during the study, sampling focused on clusters on all three campuses of UM, and later focused on sample collection at the residential halls and the UMHT for the purpose of on-campus surveillance and establishing relationships between SARS-CoV-2 levels in wastewater and the proportion of building residents/patients with documented COVID-19.
All wastewater samples were collected as grab samples (corresponding to an instant in time) using a “bottle on chain approach.” We recognize that sewage quality can vary throughout the day but due to budget limitations we chose to collect grab samples as opposed to composites which would have required the purchase and installation of autosamplers. To accommodate the variability, samples were collected at each target manhole or lift station at almost the same time each Wednesday for the purpose of eliminating, as best as possible, daily and weekly level variations.
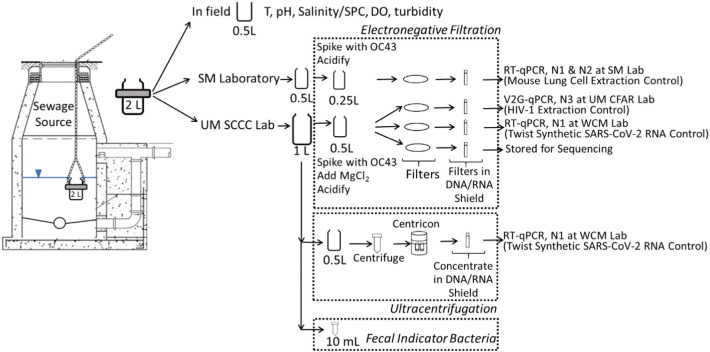
As part of sample collection, at each sampling site, a new unused 2-L bottle (HDPE) was lowered into the sewer. This 2-L sample was then split in the field into three containers: (1) a 0.5 L bottle which was sent to a commercial laboratory (Source Molecular, a LuminUltra Company, (SM)) for SARS-CoV-2 quantification; (2) a 1-L bottle which was sent to the UM SCCC laboratory for sample concentration; and (3) a 0.5 L plastic beaker which was used for basic water quality measurements in the field (temperature, pH, specific conductivity (SPC), dissolved oxygen (DO), and turbidity) using a precalibrated sonde (Xylem/YSI ProDSS) (Fig. 2 ). Details about field safety in terms of disinfection are provided in the supplemental text.
Fig. 2.
Flow diagram for sample splitting and sample concentration.
Upon receipt at the laboratory, the UM 1-L bottle was mixed. An aliquot of 10 mL was removed from the UM 1-L bottle and placed in a sterile centrifuge tube for further processing for fecal indicator bacteria by culture, as a means of confirming fecal inputs to the sampling site. E. coli was analyzed for two of the early sampling days by spread plate method on EMB agar using standard methods (Method 9221, (American Public Health Association (APHA), 2005)) to quantify colonies with characteristic green sheen. For the last eight sampling days, fecal coliform were analyzed by membrane filtration using mFC agar by culturing 1 mL and 0.1 mL aliquots from 100:1 dilutions in sterile phosphate buffered saline as per standard methods (Method 9222D, (American Public Health Association (APHA), 2005)). Colonies with characteristic blue color were quantified.
2.4. Virus concentration
Weekly samples collected in this study were concentrated via electronegative filtration by the SM laboratory and by the UM laboratory. Early during the study a subset of weekly samples were split and concentrated via ultracentrifugation. The paired samples which compared electronegative filtration and ultracentrifugation were sent to Weill Cornell Medicine (WCM) for RT-qPCR using laboratory-specific protocols described below. In addition, all weekly samples were processed for comparison between RT-qPCR (as analyzed by SM) and a new innovative technique called V2G-qPCR (as analyzed by the UM Center for AIDS Research (CFAR) laboratory), both of which used electronegative filtration for sample concentration and which utilized laboratory-specific protocols, also described below. V2G-qPCR was chosen as it has been useful in the past to detect and quantitate viral targets such as HIV-1 and Zika virus. The assay was modeled on previous work and developed early in the pandemic for the direct detection SARS-CoV-2 RNA in saliva.
Early during the WBS effort, eight samples were split and concentrated in parallel at the UM laboratory using both ultracentrifugation via Centricon-70 devices (Ahmed et al., 2020c; KWR Water Research Institute, 2020) and by electronegative filtration (Abdelzaher et al., 2008; Abdelzaher et al., 2009; Ahmed et al., 2020c; American Public Health Association (APHA), American Water Works Association (AWWA), Water Environment Federation (WEF), 2017; Bonilla et al., 2015). Splits were collected at the same timepoint and concentrated in the same laboratory at the same time. A 0.5 mL aliquot of the ultracentrifugation concentrates (1.5 mL total) were combined with 0.5 mL 2× DNA/RNA Zymo Shield. The electronegative filtration concentrates consisted of filters placed in 1.5 mL of 1× DNA/RNA Zymo Shield (Fig. 2). Sample sets of paired ultracentrifugation and electronegative filter concentrates were submitted WCM for RT-qPCR analysis. At WCM, four hundred microliters of each wastewater concentrate was transferred to a deep-well plate for total nucleic acid (TNA) extraction. All samples were extracted on the Tecan DreamPrep Fluent 480 automated workstation using the Zymo Research DNA/RNA Viral Magbead kit to produce 60 μL of purified DNA/RNA. Viral abundance of SARS-CoV-2 was determined by RT-qPCR using the Applied Biosystems QuantStudio 6 Flex using a one-step TaqMan Fast Virus Master Mix which targets the N1 gene (Centers for Disease Control and Prevention (CDC), 2020). Twist Synthetic SARS-CoV-2 RNA Control 2 (MN908947.3) was used as a control, which comes as 106 copies per μL. Dilution of the synthetic SARS-CoV-2 standard brought copies down to 102 per reaction before use. Samples were run in triplicate, with duplicate controls.
To facilitate the comparison between RT-qPCR and V2G-qPCR, the 2-L sewage grab sample was split with the 0.5 L bottle going to the SM commercial laboratory, and the 1-L bottle going to the UM SCCC laboratory. Concentration for both samples was done by electronegative filtration in the corresponding separate labs (Fig. 2). Wastewater was filtered the same day as collected for all UM samples, and also the same day (except for once during all the days of sampling) for the SM samples. To prepare samples for electronegative filtration, a known number of betacoronavirus OC43 particles were added prior to sample processing to assess recovery efficiency. For SM, a high concentration (about 107 copies) of heat inactivated (at 60 °C for 60 min) human coronavirus HCoV-OC43 (ZeptoMetrix) was spiked into the sample volume to be filtered as a virus recovery control. For each filtration batch, an additional OC43 positive control sample was included which contained the same amount of heat-inactivated OC43 added to the lysis buffer. All samples (n = 95) included the OC43 process control spike for SM. For the UM laboratory samples the OC43 process control was grown in cell culture using HCT-8 or Vero cells (ATCC) and quantified in triplicate using Volcano Second Generation qPCR (V2G-qPCR) (details of these assays are provided below). Quantitative OC43 spikes (heat inactivated at 56 °C for 15 min) were added between 105 and 106 gc/L for the UM samples. For the UM samples, the last 51 samples (starting on November 11th) included the process control spike. After addition of the process control, samples were then adjusted by acidification. The SM samples were acidified to a pH of 3.5 using 2 N HCl. For the UM laboratory samples, MgCl2 was added to a concentration of 50 mM and acid (10% HCl) was added drop by drop to a target pH between 4.5 and 3.5, as per standard methods (Method 9510, (American Public Health Association (APHA), American Water Works Association (AWWA), Water Environment Federation (WEF), 2017)). Once the samples were prepared, they were mixed and filtered through an electronegative membrane (0.45 μm pore size, 47 mm diameter, SM: Pall HA/MCE membrane, UM: Millipore HAWP4700) to clogging (from 20 mL to 200 mL). The filter prepared by SM was placed in a 2 mL bead tube containing 700 μL of PM1 lysis buffer (Qiagen AllPrep PowerViral DNA/RNA Kit) with beta-mercaptoethanol and stored at -20 °C until DNA/RNA extraction which was completed within 7 days of freezing. Three filters were prepared per sample at the UM laboratory. Each UM filter was folded and placed into its own tube (5 mL Eppendorf tube for V2G-qPCR analysis or a 2 mL sterile tube for all other analyses). Tubes contained 1.5 mL of DNA/RNA Shield (Zymo) forming a sewage concentrate that was stored at 4 °C until analysis.
2.5. SARS-CoV-2 quantification for comparison of weekly samples
Among the three UM filters, one was processed for rapid analysis by the innovative V2G-qPCR method developed at the UM CFAR. One of the other two UM SCCC filters was sent to WCM and the other was stored for later analyses. The RNA from the UM filter analyzed by V2G-qPCR was extracted using a Zymo Quick-RNA Viral Kit using either 500 μL (first 10 sampling days) or 250 μL (last 2 sampling days) of the sewage concentrate to recover purified RNA (30 μL). The volume of concentrate was reduced from 500 to 250 μL to address inhibition, especially for highly turbid (>100 ntu) samples. Primers and reporter probe for the V2G assay bind to the same region as the nucleocapsid N3 target (Centers for Disease Control and Prevention (CDC), 2020) but differ substantially. Reagent sequences were based on GenBank accession number NC_045512 and corresponded to 28645TGCTAACAAAGACGGCATCA (forward), 28751GTAGCACGATTGCAGCATTG (reverse) and 56-FAM/ACATTGGCA/ZEN/CCCGCAATCCTGCT3IABkFQ (probe).
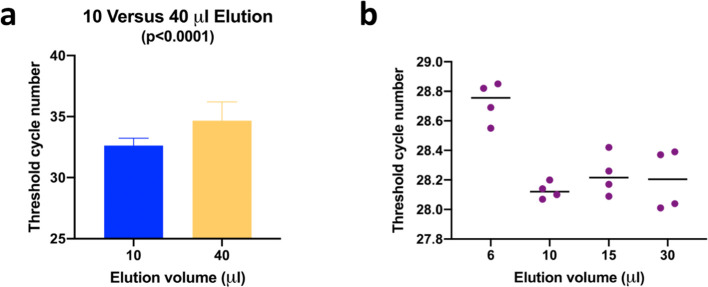
The V2G-qPCR method uses a novel polymerase capable of reading both RNA and DNA templates and therefore does not require a separate cDNA synthesis step (Blatter et al., 2013). Aliquots of purified RNA were used in singleplex reactions to quantitate up to four targets using a BioRad CFX Connect real-time instrument and FAM reporter dye-labeled probes. Targets included the SARS-CoV-2 N3 nucleocapsid gene (as modified from Lu et al., 2020b) (5 μL of the RNA concentrate), the human-specific general house-keeping gene B2M (M.Sharkey, unpublished) (2 μL), the process control OC43 (3 μL), and HIV-1 RNA (M.Sharkey, unpublished) as the inhibition control (2 μL). The B2M target is commonly used as an endogenous control to normalize relative gene expression levels by RT-qPCR. Both SARS-CoV-2 and B2M targets were analyzed for all sampling days. The OC43 target was analyzed for samples collected during the last 6 sampling days (starting November 11th) and the HIV-1 target was analyzed for samples collected during the last 4 sampling days (starting November 25th) to measure the degree to which PCR amplification was inhibited by wastewater components that co-purified with RNA. Early during the study, to minimize PCR inhibition by dilution effect, we doubled the amount of water to elute RNA from the purification columns. Unexpectedly, increasing the volume of elution water led to a more pronounced inhibition of the PCR amplification reaction that was reduced by eluting with a smaller volume of water (Fig. 3a). Ct values were determined in duplicate for twelve sample sites using final RNA elution volumes of 10 or 40 μL. Comparison of the means of the two data sets showed an average shift of two cycles when the larger elution volume was used which was statistically significant (p < 0.0001). Recovery of RNA was compared using different elution volumes and it was determined that 10 μL was sufficient to elute the RNA without losses (Fig. 3b). We hypothesize that the reason for this observation is that a larger volume flushes more inhibitory material through the column matrix. Using a minimal volume hydrates the silica such that the RNA is efficiently released, while inhibitory substances are retained on the silica bed.
Fig. 3.
RNA column elution volume significantly affects target PCR amplification efficiency. PCR inhibition is more pronounced when using an elution volume of 40 μL versus 10 μL (panel a). RNA elution from Zymo columns is efficient when using 10 μL water (panel b).
For SM, all filters were extracted using AllPrep PowerViral DNA/RNA Kit (Qiagen) bead beating kit to produce 70 μL of purified DNA/RNA. In addition, 5 ng of Mouse Lung Total RNA (Takara Bio USA) was added into the sample lysate to evaluate extraction efficiency and PCR inhibition. The percentage lysate recovered during extraction was also considered as part of the evaluation of inhibition. RT-qPCR analysis was performed on the StepOne Plus system (Thermo Fisher Applied Biosystems), and Taqman Fast Virus 1-Step Master Mix (Thermo Fisher) was used. Five microliters of DNA/RNA template were added into a final volume of a 20 μL RT-qPCR reaction. Each sample was analyzed in duplicate. For quantification of SARS-CoV-2 RNA, the N1 and N2 primer and probe assays (Centers for Disease Control and Prevention (CDC), 2020) were employed. Synthetic RNA standard (ATCC, VR-3276SD) was diluted to create the standard curve for N1 and N2. For relative quantification of Mouse ACTB, Mouse ACTB endogenous control primers and probe set (FAM reporter, primers unlimited, ThermoFisher) was used. Mouse Lung RNA (2.5 ng) was run as duplicates with each extraction batch as a reference. For quantification of OC43, published OC43 primers and probe (Dare et al., 2007) were used. OC43 genomic RNA standard (ATCC, VR-1558DQ) was diluted to create the standard curve. After RT-qPCR and qPCR runs, data was processed on StepOne Software (v2.3). Ct values from qPCR measurements were converted to genomic copies (gc) per reaction using standard calibration curves of known levels of known standards. The gc per reaction were then converted to gc/L of original sewage sample using mass balance considerations. The results from SM were used to check the results from the V2G-qPCR analyses, retrospectively.
2.6. Statistical analysis
Wastewater results were imported into Excel (Microsoft Office 365 Pro Plus). Correlations between the RT-qPCR results and V2G-qPCR were evaluated using Pearson correlation coefficients (R2) using base-10 logarithm-transformed values. Correlations were considered strong for R2 greater than 0.5 (Shibata et al., 2004) and were considered significant for p values less than 0.05. Statistical differences between means of wastewater data were evaluated using single factor ANOVA. For sets of analyses showing statistical differences, student t-tests (assuming paired two samples for means, two tailed with alpha at 0.05) were used to evaluate differences between two specific data sets. Statistical differences between the variance of data sets were analyzed using the F-test with an alpha value of 0.05.
Human surveillance data were processed by assuming that subjects testing positive for COVID-19 on a given day were infected many days before and were shedding the virus since contracting it. Therefore, this shedding of the virus can be traced in the wastewater samples even before their clinical diagnoses. Thus, in our analysis we computed cumulative and daily lag-specific COVID-19 positivity rates 7 days pre- (or -7, wastewater signal before human cases) consistent with early WBS studies (Medema et al., 2020a) and 15 days post (+15, wastewater signal after human cases) human case date which is consistent with the duration of illness (Wang et al., 2020). We computed cumulative positivity rate by dividing the total number of positive cases up to a given lag divided by the total number of tests performed up to that lag. For example, to compute the -4 day lag the cumulative positivity rate can be computed as the sum of COVID-19 positive cases from 7th day through 4th day before the wastewater sampling date divided by the sum of total tests performed during the same time period, and coefficient was multiplied by 1000. The cumulative l th day lagged positivity rate for a given time (t) can be expressed as y t±l ~ 1000 ([Σlϵ( − 7,15) Ct±l] / [Σlϵ( − 7,15) Tt±l]). Where Ct±l is the COVID-19 positive cases on l th lag from time t (i.e., day of wastewater sampling). The daily lag-specific positivity rate was computed as the ratio of the number of positive cases on l th day after a given date divided by the total tests on the corresponding lagged day and multiplied by 1000. The time-lagged positivity rates were analyzed with respect to SARS-CoV-2 RNA concentration found in the wastewater samples using descriptive statistics and location-specific linear regressions with random effect, since the pattern of COVID-19 positivity rate is likely to have unique pattern for units draining to a given wastewater sampling location. Since the SARS-CoV-2 RNA concentration (gc/L) was positively skewed, it was log transformed. In the descriptive analysis, RNA concentration was categorized into four groups: below detection limit, less 1 K, 1 K to 10 K and 10 K+.
Human surveillance data were available for student residents on campus, for student non-residents who visit campus for hybrid courses, and for faculty/staff. The lag correlation analysis described above was performed using the student resident data given that this population was on campus full-time on a daily basis.
3. Results and discussion
3.1. Water quality
Overall, water quality results (Fig. S-1) showed that each sewer had unique characteristics that could potentially influence the quantity and persistence of SARS-CoV-2 (Ahmed et al., 2020d; Buonerba et al., 2021; La Rosa et al., 2020). The pH values per site ranged from 6.9 to 8.8, the average SPC varied from 149 μS/cm to 3420 μS/cm, and the average turbidity of the wastewater by site varied from 1.5 ntu to 161 ntu (Fig. S-1b,c,e). Of interest was that dissolved oxygen levels were higher than expected with average values per sampling location ranging from 4.0 to 7.7 mg/L (Fig. S-1d) with values at saturation for some sampling events. Among the water quality parameters measured, temperature (with average values per sampling location ranging from 22.0 to 32.6 °C, Fig. S-1b) is known to impact the persistence of the SARS-CoV-2 signal with higher temperature resulting in higher viral RNA degradation rates (Hart and Halden, 2020; Ma et al., 2020; Nazari Harmooshi et al., 2020; Carducci et al., 2020; Hokajärvi et al., 2021). Different sites showed statistical differences in mean and variances in water quality parameters. In general, buildings showed higher variability in water quality compared to clusters. Statistical details are further described in the supplemental text.
Correlations were evaluated between water quality and SARS-Cov-2 levels. All correlations were weak (R2 < 0.1) and insignificant. Although significant differences were observed in water quality parameters between sites, these measures did not correlate with the SARS-CoV-2 RNA signal.
We also found the variations in bacteria levels to be surprising. At wastewater treatment plants the typical level of E. coli in sewage (at the community scale) is 10,000 colony forming units (CFU) per mL of wastewater (Roca et al., 2019). Some sewage samples showed these levels but there was also sewage from clusters that were at levels that were much less, less than 10 CFU per mL. As a result, the fecal indicator bacteria analyses were switched to measurements of fecal coliform by membrane filtration to confirm whether in fact the levels of fecal bacteria were low. Again, with the fecal coliform plates we found that levels in the sewage were as expected for some sites, on the order of 10,000 per mL. But there was sewage from some buildings and clusters that were consistently low. This was confirmed with two different sets of agar plates and was observed repeatedly on a weekly basis. These clean plates were likely associated with low levels of feces in the wastewater at the time of sampling and the likely effects of chlorine residual from the tap water. This resulted in a change in our protocol where samples were processed with the same procedures used to collect drinking water. A reductant, sodium thiosulfate, was added to the sample collection bottles (0.1 g added per liter of wastewater) starting November 25th to reduce the chlorine residual. Even with the reductant added clean agar plates were still observed for some sites.
3.2. Virus concentration
Ultracentrifugation using Centricon-70 devices and electronegative filtration performed similarly for the detection of SARS-CoV-2. Specifically, the mean Ct value for the ultracentrifugation concentrates (29.2) was higher than the mean Ct value for the electronegative filter concentrates (26.9). The Ct mean values were not statistically different at 95% confidence (p = 0.097) and consistent with other studies that have compared sample concentration methods (Lu et al., 2020a; Pecson et al., 2021). The equivalent volume per concentrate distributed to WCM was 16 and 17 mL for ultracentrifugation and 50 to 140 mL for electronegative filtration resulting in a factor of 3 to 8 times more original wastewater sample volume for qPCR reactions using electronegative filtration. The larger equivalent volume of wastewater analyzed using electronegative filtration likely contributed to the lower Ct values on average.
In addition to showing comparable results between both ultracentrifugation and electronegative filtration concentration methods, we found that the supplies for electronegative filtration were more readily available. The nature of the electronegative filtration process also allowed for the preparation of multiple filters from the same sample without sacrificing concentration factors. As a result, sample concentration by ultracentrifugation was dropped early during the study and electronegative filtration was run for all samples.
3.3. Comparison of V2G-qPCR and RT-qPCR results
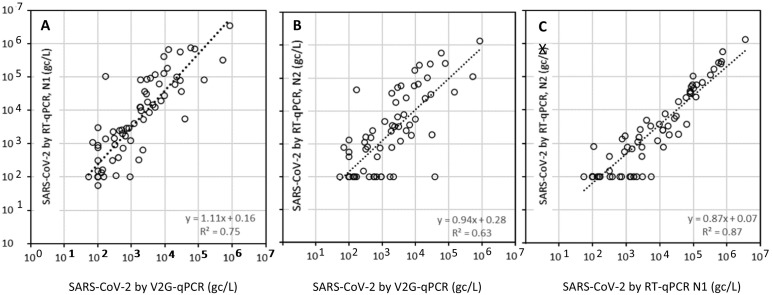
The overall comparison between the UM CFAR V2G-qPCR method and the SM RT-qPCR method shows a strong correlation between the two methods with N1 RT-qPCR detection showing a slightly stronger correlation (R2 = 0.75, p < 0.01) in comparison to N2 RT-qPCR (R2 = 0.63, p < 0.01) (Fig. 4 ). The correlations between N1 and N2 RT-qPCR detection (R2 = 0.87, p < 0.01) were also, as expected, strong. Correlations between V2G-qPCR and RT-qPCR results were particularly good considering that two different laboratories were used for sample concentration and considering the different amplification processes, detection technologies, and target genes. On average the results from the N1 RT-qPCR method were about 10% higher than those of the V2G-qPCR method, whereas results from the N2 RT-qPCR method were about 6% lower than those of the V2G-qPCR method. The detection limits for both the V2G-qPCR and RT-qPCR technologies were similar, on the order of a few 100's of gc/L. SM reports a limit of detection (LOD) of 3 copies per reaction and a limit of quantitation (LOQ) of 10 copies per reaction. Assuming a 95% extraction efficiency of 5 μL RNA input in the qPCR reaction, 70 μL of total RNA extract, and 200 mL filtration volume, then the LOD is estimated as 220 gc/L and the LOQ is estimated as 740 gc/L. Similarly for V2G-qPCR the LOD is estimated at 1 copy per reaction through validation with standards, and the LOQ at 10 copies per reaction. Assuming a 95% extraction efficiency of 5 μL RNA input in the qPCR reaction, 30 μL of total RNA extract, and 100 mL filtration volume, then the LOD is estimated as 70 gc/L and the LOQ is estimated as 700 gc/L.
Fig. 4.
Comparison between SARS-CoV-2 RNA levels (gc/L) between V2G-qPCR and RT-qPCR detecting the N1 gene (panel A), V2G-qPCR detecting a modified N3 gene and RT-qPCR detecting the N2 gene (panel B), and RT-qPCR detecting the N1 gene and the N2 gene (panel C).
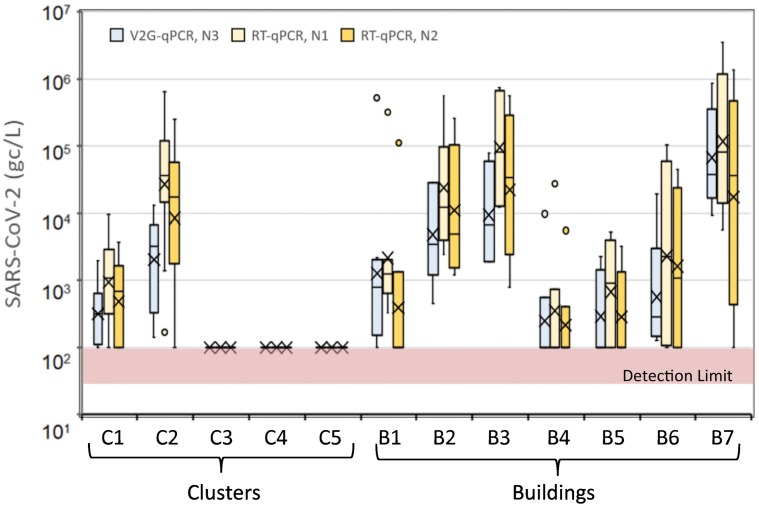
When comparing results for individual sites, the sites that detected consistently below detection limits (C3, C4, and C5) showed below detection limit values for both the V2G-qPCR and RT-qPCR method (Fig. 5 ). For the remaining sites, the N1 RT-qPCR results and the N2 RT-qPCR results were statistically not different as expected given that these were processed from the same concentrate and at the same laboratory. Of particular interest is that for all sites, V2G-qPCR and N2 RT-qPCR results were also statistically not different (p > 0.05), which is particularly promising given that different laboratories and methods were used for quantification of SARS-CoV-2. The only statistical differences observed between V2G-qPCR and N1 RT-qPCR were for a subset of the sites: for C1 (p = 0.049) and C2 (p = 0.01). For all other sites, V2G-qpcr and N1 RT-qPCR results were not statistically different.
Fig. 5.
Ranges of SARS-CoV-2 measurements for different clusters and buildings as determined by V2G-qPCR (left box plot, light blue) and by RT-qPCR (right box, light orange for N1 gene and darker orange for N2 gene).
To enable rapid turn-around time, V2G-qPCR results were used for UM SARS-CoV-2 wastewater surveillance. Analysis results were reported to the university administration leadership within 24 h of sample collection, to inform decision-making on human health surveillance and disease prevalence mitigation measures. The main advantage of the V2G-qPCR method is that is it simpler than the traditional RT-qPCR methods in that it does not require a separate cDNA synthesis step which reduces assay time and cost. Due to genetic modifications, V2G polymerase activity is robust when using crude cell lysates (Chovancova et al., 2017) and when amplifying RNA from unprocessed biological fluids, such as saliva and urine. Similar, robust amplification efficiency has been observed in this study when using RNA purified from wastewater concentrates with optimal column elution volumes. The standard turn-around time from receipt of the sewage concentrates to SARS-CoV-2 quantitation was 2.5 h. The reduced time for analysis is facilitated by a 50-minute cycling time for V2G-qPCR in comparison to a 1.5 to 2 h cycling time for most RT-qPCR assays. Although results were routinely available within 24 h of sample collection, a 12-hour turn-around time was possible and was achieved for V2G-qPCR when needed.
3.4. Trends observed for SARS-CoV-2 in wastewater
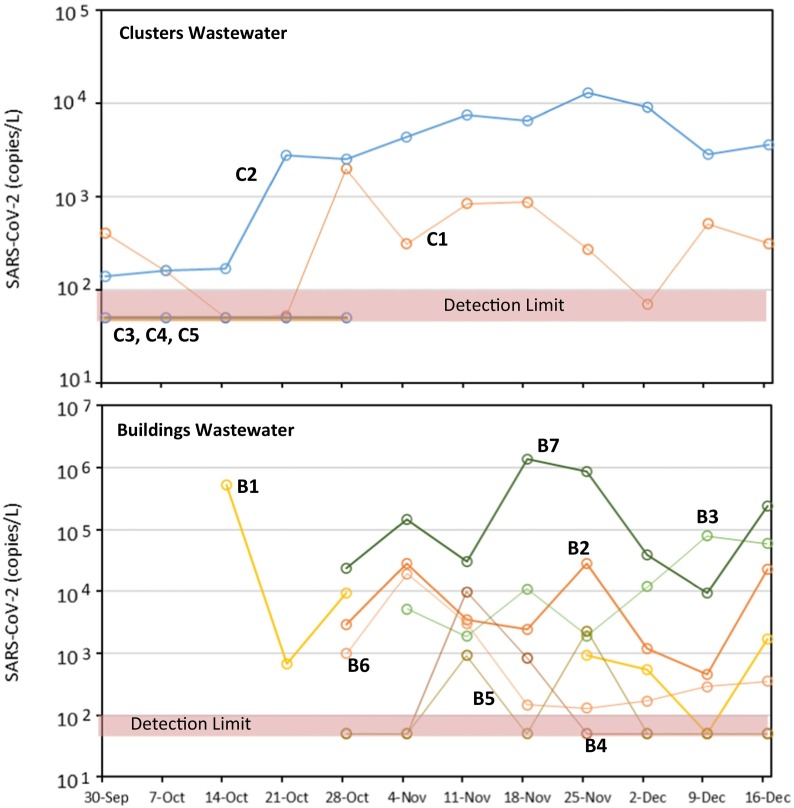
A wide range of SARS-CoV-2 virus concentrations were observed in wastewater (Fig. 6 ). SARS-CoV-2 levels in wastewater for some clusters (C3, C4, and C5) were consistently below detection. These clusters had warmer water and one cluster (C3) also had high salinity. However, wastewater from cluster C2 was consistently at high levels with all factors consistent with water conducive to higher SARS-CoV-2 RNA levels (it had a lower water temperature, near neutral pH, low salinity, and moderate turbidity). On average, C2 viral levels were in the 103 to 104 gc/L range. Wastewater virus concentrations from individual buildings were also variable. For example, wastewater from building B4 and B5 were generally below detection limits for SARS-CoV-2 RNA. Wastewater from some of the other buildings (e.g., B1 and B7) showed very high viral load levels, as high as 106 gc/L.
Fig. 6.
Time series of SARS-CoV-2 via V2G-qPCR in wastewater from clusters of buildings and individual buildings during the September to December 2020 sampling period.
As seen in Fig. 5, for virus concentration levels measured using the V2G-qPCR method, clusters C3, C4, and C5 had statistically lower SARS-CoV-2 levels relative to clusters C1 and C2 and relative to buildings B2, B3, and B7 (all p < 0.01). SARS-CoV-2 levels for B7 were statistically different than levels observed at all clusters (C1 through C5, p < 0.01) and all other buildings (B1, p = 0.02; B2, p = 0.03; B4, p = 0.04; B5, p < 0.01; B6, p < 0.01) with the exception of B3 (p = 0.07). Except for the extremely low variances for C3, C4, and C5 due to values below detection limits, none of the variances among the remaining sampling locations were statistically different.
For levels measured using RT-qPCR, clusters C3, C4, and C5 had statistically lower SARS-CoV-2 levels relative to all other sites. Based upon N1 gene quantification, the means of C1, B4, and B5 were statistically less than C2, B2, B3, and B7 (p < 0.01). Results were similar based upon N2 gene quantification with the means of C1, B4, and B5 statistically less than C2, B2, and B3 (p < 0.01) with the exception of B7 which was not statistically different due to the higher variation of the N2 values for this site. The variance of B7 was not statistically different from other sites except for C1 (p < 0.01) and B4 (p = 0.03).
Over time (Fig. 6), results show that the variability of the wastewater from clusters was more gradual. Wastewater from individual buildings had higher variability, and generally gave a strong positive or negative signal. For example, the wastewater from building B4 had a very interesting trend. The wastewater for this building started at below detection limits, went up to the 104 gc/L, then one week later dropped to 103 gc/L, and then the week after fell to below detection limits again. Apparently a source of SARS-CoV-2 was present in the November 11th time frame in building B4 releasing SARS-CoV-2 RNA into the sewer. The wastewater from building B5 showed similar trends with values going above and below detection limits. The wastewater from buildings B2 and B3 were more constant in the 103 to 104 gc/L range. In addition, wastewater from building B1 and B7 varied from 103 to 106 gc/L during this time frame.
3.5. Correlation of wastewater SARS-CoV-2 RNA with COVID-19 human cases
In regards to human health surveillance during this period (August to December 2020), 536 of the 27,526 student resident participants tested were positive for COVID-19, reflecting an average positivity rate of 194 cases per 1000 tested participants. The daily average positivity rate was 23.6 cases per 1000 tested subjects (95% confidence interval (CI) = 18.1 to 29.2 cases/1000 tests). Of note, this positivity rate was lower than the rate in the greater Miami-Dade County community.
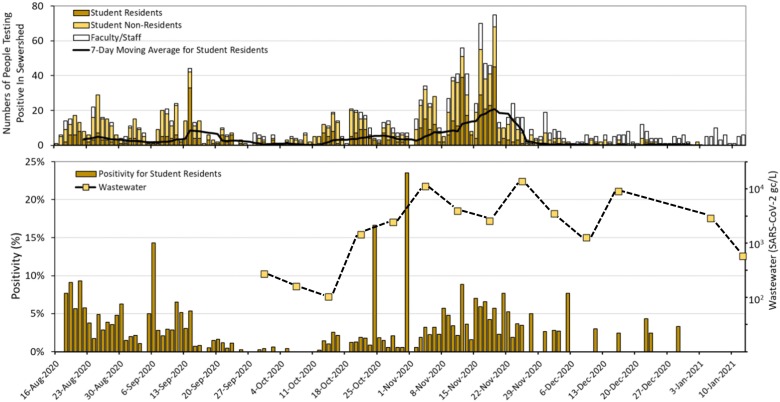
To increase statistical power, human health data were aggregated for the entire campus with wastewater results represented by sites C1 and C2 and building B2 which was outside of the C1 and C2 sewer sheds. Of note C2 received wastewater from B3, B4, B5, and B7. C1 received wastewater from B6. Building B1 is on a separate campus whose human population was not included in the aggregation. From 38 wastewater samples of three aggregate sites, only two were below the detection limit. The average SARS-CoV-2 concentration was 1283 gc/L (95% CI = 696 to 2364.8 RNA copies/L). A temporal comparison of human surveillance with the wastewater samples suggests that the concentration of SARS-CoV-2 RNA in wastewater sample was followed by spikes in the COVID-19 positivity rates, especially toward the end of October 2020 (Fig. 7 ).
Fig. 7.
Temporal distribution of COVID-19 positive cases for student residents, student non-residents, and faculty/staff plus 7-day moving average for student residents (panel A) and positivity rates (panel B) among student residents and SARS-CoV-2 levels in wastewater (log scale on secondary y axis). Wastewater levels of SARS-CoV-2 represent the average of C1, C2 and B2.
In the time-lagged analyses, COVID-19 positivity rate increased as SARS-CoV-2 RNA concentration increased (Fig. 8 ). In the daily lag-specific analysis, −5 through −3 day and 3 through 5 day lagged positivity rates were significantly associated with SARS-CoV-2 RNA concentrations in wastewater samples (Table S-1). The association of SARS-CoV-2 RNA concentration in the wastewater was strongest with the 3rd day lagged positivity rate (β ~ 8.99; 95% CI = 0.90 - 17.08; p < 0.05), followed by the -4th day lagged positivity rate (β ~ 8.68; 95% CI 9.30–70.64; p < 0.05). This suggests that most COVID-19 positive cases peaked four days after and three days before high concentrations of SARS-CoV-2 RNA were found in the wastewater samples. On average 100 copies of the SARS-CoV-2 RNA were associated with a positivity rate (numbers per 1000 tested) of 40 and 41 on 4th day after and the third day before the wastewater sampling date (40–8.68 × ln(100); 41–8.99 × ln(100)), respectively.
Fig. 8.
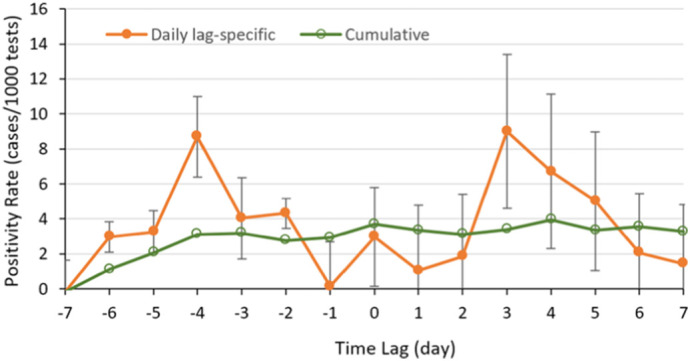
Time-lagged positivity rate with respect to log (SARS-CoV-2 RNA concentration in gc/L) in the wastewater samples between October 30, 2020 and January 2, 2021. The cumulative model included positivity rates between 7 day and a given day before/after the wastewater sampling day. In the daily-lag specific model, positivity rate was computed for a given day before/after the wastewater sampling day. Error bars correspond to 95% confidence limits on positivity rate.
When analyzing cumulative time-lagged analysis, the -6th through 0 day lagged cumulative COVID-19 positivity rates showed significant association with the virus RNA concentration in the wastewater samples (Table S-1). This association was not significant from 1 through 5th day lag and was also significant on the 6th day lag. There were differences in the association of time-lagged COVID-19 positivity rates with the virus RNA in the wastewater samples when disaggregating by C1, C2, and B2. The strongest associations were observed in C2 followed by B2.
4. Conclusions
We report here on lessons learned from establishing a wastewater monitoring program to supplement human surveillance of COVID-19. From this study we observed that SARS-CoV-2 RNA concentrations in individual buildings were more variable than in clusters of buildings. Concentrations at building and cluster scales varied by orders-of-magnitude from detection limits of 101 through 106 gc/L allowing for the log transformation of the data to observe trends. In terms of overall quality of the wastewater, we observed that basic physical-chemical parameters were influenced by the water source but water quality parameters were not correlated with SARS-CoV-2 levels. Additionally, the potable water source that services the building is chlorinated for disinfection, the residual of which is believed to reduce bacteria in the wastewater to levels that were lower than expected.
Ultracentrifugation and electronegative filtration concentration methods were found to be comparable. Electronegative filtration was facilitated by the ease of obtaining supplies and provided for efficient sample splitting allowing for multiple filters to be prepared and shared among laboratories without sacrificing processing volume.
We also learned that new innovative technologies, such as V2G-qPCR, can simplify the detection and quantification of SARS-CoV-2 in wastewater. The approach was initially meant to serve as an interesting comparator for the more mainstream RT-qPCR approaches but developed into a reliable and consistent assay for quantifying SARS-CoV-2 RNA purified from wastewater. Since RNA extracted from wastewater is amplified directly with V2G-qPCR, the cDNA synthesis step is bypassed, reducing both assay time and cost. Using this new tool, it was possible to have results from the start of sampling to the end of detection within 12 h, which is fast compared to standard techniques, and therefore can be useful for an early detection system for COVID-19.
Comparison of wastewater results against human surveillance data suggests that SARS-CoV-2 measures in wastewater can provide an early warning of impending COVID-19 outbreaks. Results showed that high concentration of SARS-CoV-2 RNA in wastewater samples on a given day indicates undetected COVID-19 cases. These undetected cases will likely be observed 4 days after the observed increase in wastewater RNA levels. To fine tune the optimum lag time, wastewater sampling at a higher frequency (daily as opposed to weekly) would facilitate one-to-one matching of COVID-19 positivity rates with measurements of SARS-CoV-2 levels in wastewater. It is possible that with daily sampling, the early warning period could be longer than 4 days allowing more time to identify positive subjects and thereby possibly reducing disease transmission.
This ongoing study required the integration of field sampling information, concentration information, and detection results from multiple laboratories that use different technologies and processing protocols. Although detailed metadata were recorded at each step of sample processing, virus quantification, and data analysis, the specific metadata parameters and descriptions were not standardized prior to the start of the project. A considerable effort was therefore spent to manually combine and harmonize the various sources of information to report the results of this study. To continue and expand SARS-CoV-2 wastewater surveillance studies and to integrate these results with those of other studies to detect SARS-CoV-2 or other wastewater datasets requires a systematic approach to metadata standardization and data harmonization (Stathias et al., 2018; Vempati et al., 2014). We anticipate that sample processing and analysis methods which integrate metadata standardization and data harmonization such as those utilized in high throughput clinical labs can be tailored for use as part of intense wastewater sampling programs used to gauge the health of a community.
Overall, this study showed that the challenges associated with tracking disease outbreaks associated with the COVID-19 pandemic can be met through a multi-pronged approach that integrates comprehensive human surveillance of the disease with environmental surveillance of the virus. In the case of COVID-19, the RNA of the etiologic agent of disease, SARS-CoV-2, was found to unexpectedly be excreted in urine and feces of both symptomatic and asymptomatic people. Although COVID-19 is a respiratory disease, and it would be expected in respiratory fluids, it also has been found in wastewater, allowing for an alternative approach to detecting early onset of outbreaks by measuring markers of the pathogen in wastewater. Future work should focus on expanding techniques and protocols for environmental monitoring of infectious agents for the purpose of tracking disease outbreaks.
CRediT authorship contribution statement
Mark Sharkey: Conceptualization, Methodology, Visualization, Formal Analysis, Writing – Original Draft. Naresh Kumar: Conceptualization, Methodology, Visualization, Formal Analysis, Writing – Original Draft. Alejandro Mantero: Conceptualization, Methodology, Visualization, Formal Analysis, Writing – Original Draft. Kristina M. Babler: Conceptualization, Methodology. Melinda Boone: Conceptualization, Methodology. Yoslayma Cardentey: Methodology. Elena Cortizas: Conceptualization, Methodology. George Grills: Conceptualization, Methodology, Writing – Review, Funding Acquisition. James Herrin: Conceptualization, Methodology, Writing – Review. Jenny Kemper: Conceptualization, Methodology. Richard Kenney: Conceptualization, Methodology. Erin Kobetz: Conceptualization, Methodology, Supervision. Jennifer Laine: Conceptualization, Methodology, Supervision. Walter Lamar: Conceptualization, Methodology, Writing – Review, Supervision. Christopher Mason: Conceptualization, Methodology, Supervision, Funding Acquisition. Anda Quintero: Conceptualization, Methodology, Writing – Review. Brian Reding: Conceptualization, Methodology. Matthew Roca: Methodology. Krista Ryon: Conceptualization, Methodology, Writing – Review. Stephan Schürer: Conceptualization, Methodology, Writing – Review, Supervision, Funding Acquisition. Bhavarth Shukla: Conceptualization, Methodology, Writing – Review, Supervision. Natasha Solle: Conceptualization, Methodology, Supervision. Mario Stevenson: Resources. Thomas Stone: Methodology. John Tallon: Conceptualization, Methodology, Resources. Sreeharsha Venkatapuram: Graphics. Dusica Vidovic: Conceptualization, Methodology. Sion Williams: Conceptualization, Methodology. Benjamin Young: Conceptualization, Methodology, Formal Analysis, Writing – Review. Helena Solo-Gabriele: Conceptualization, Methodology, Visualization, Formal Analysis, Writing – Original Draft, Supervision, Project Administration, Funding Acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported financially by the University of Miami (Coral Gables, FL) administration, with in-kind contributions from University Facilities, University Environmental Health and Safety, and University of Miami Health Safety Division. Laboratory facilities and support were made available in-kind through the Sylvester Comprehensive Cancer Center, the Miami Center for AIDS Research, and the Miami Clinical and Translational Science Institute. Later parts of the research reported in this publication were supported by the National Institute On Drug Abuse of the National Institutes of Health (NIH) under Award Number U01DA053941. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We are also thankful to our many colleagues with whom we have interacted and discussed best practices for wastewater sample collection, concentration, and analysis for SARS-CoV-2.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.149177.
Appendix A. Supplementary data
Supplementary material
References
- Abdelzaher A.M., Solo-Gabriele H.M., Wright M.E., Palmer C.J. Sequential concentration of bacteria and viruses from marine waters using a dual membrane system. J. Environ. Qual. 2008;37:1648–1655. doi: 10.2134/jeq2007.0238. [DOI] [PubMed] [Google Scholar]
- Abdelzaher A.M., Solo-Gabriele H.M., Palmer C.J., Scott T.M. Simultaneous concentration of enterococci and coliphage from marine waters using a dual layer filtration system. J. Environ. Qual. 2009;38:2468–2473. doi: 10.2134/jeq2008.0488. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passengers aircraft and cruise wastewater: a surveillance tool for assessing the presence of COVID-19 infected travelers. J. Travel Med. 2020;728 doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-QPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;18:739. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Public Health Association (APHA) American Water Works Association A.W.W.A., Water Environment Federation (WEF) 21st edition. APHA, AWWA, WEF; Washington DC: 2005. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- American Public Health Association (APHA), American Water Works Association (AWWA), Water Environment Federation (WEF) 9510 Detection of Enteric Viruses. 23rd edition. APHA, AWWA, WEF; Washington DC: 2017. Standard methods for the examination of water and wastewater. [Google Scholar]
- Bannwarth A., Morelato M., Benaglia L., Been F., Esseiva P., Roux C., Delemont O. The use of wastewater analysis in forensic intelligence: drug consumption comparison between Sydney and different European cities. Forensic Sci. Res. 2019;4(2):141–151. doi: 10.1080/20961790.2018.1500082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanot D., Singh T., Verma S.K., Sharad S. Stigma and discrimination during COVID-19 pandemic. Front. Public Health. 2021;8:829. doi: 10.3389/fpubh.2020.577018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., De Los Reyes F.L., Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., et al. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(13):7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Blatter N., Bergen K., Nolte O., Welte W., Diederichs K., Mayer J., Wieland M., Marx A. Structure and function of an RNA-reading thermostable DNA polymerase. Angew. Chem. Int. Ed. Engl. 2013;52(45):11935–11939. doi: 10.1002/anie.201306655. [DOI] [PubMed] [Google Scholar]
- Bonilla J.A., Bonilla T.D., Abdelzaher A.M., Scott T.M., Lukasik J., Solo-Gabriele H.M., Palmer C.J. Quantification of protozoa and viruses from small water volumes. Int. J. Environ. Res. Public Health. 2015;12:7118–7132. doi: 10.3390/ijerph120707118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer A.F., Eisenberg J.N.S., Pomeroy C.D., Shulman L.M., Hindiyeh M., Manor Y., Grotto I., Koopman J.S., Eisenberg M.C. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc. Natl. Acad. Sci. U. S. A. 2018;115(45):E10625–E10633. doi: 10.1073/pnas.1808798115. https://www.scopus.com/inward/record.uri?eid=2-s2.0- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonerba A., Corpuz A.V.A., Ballesteros F., Choo K.-H., Hasan S.W., Korshin G.V., Belgiorno V., Barceló D., Naddeo V. Coronavirus in water media: analysis, fate, disinfection and epidemiological applications. J. Hazard. Mater. 2021;415 doi: 10.1016/j.jhazmat.2021.125580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci A., Federigi I., Liu D., Thompson J.R., Verani M. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes, June 6. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
- Centazzo N., Frederick B.-M., Jacox A., Cheng S.-Y., Concheiro-Guisan M. Wastewater analysis for nicotine, cocaine, amphetamines, opioids and cannabis in New York City. Forensic Sci. Res. 2019;4(2):152–167. doi: 10.1080/20961790.2019.1609388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovancova P., Merk V., Marx A., Leist M., Kranaster R. Reverse-transcription quantitative PCR directly from cells without RNA extraction and without isothermal reverse-transcription: a ‘zero-step’ RT-qPCR protocol. Biol. Methods Protoc. 2017;2(1):bpx008. doi: 10.1093/biomethods/bpx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dare R.K., Fry A.M., Chittaganpitch M., Sawanpanyalert P., Olsen S.J., Erdman D.D. Human coronavirus infections in rural Thailand: a comprehensive study using real-time reverse-transcription polymerase chain reaction assays. J. Infect. Dis. 2007;196(9):1321–1328. doi: 10.1086/521308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total Environ. 2020;726 doi: 10.1016/j.scitotenv.2020.138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda M., Mustafa F., Maraqa M., Rizvi T., Aly Hassan A. Wastewater surveillance for SARS-CoV-2: lessons learnt from recent studies to define future applications. Sci. Total Environ. 2021;10 doi: 10.1016/j.scitotenv.2020.143493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020:737. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020:730. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokajärvi A.-M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.-M., Kankaanpää A., Gunnar T., Al-Hello H., Blomqvist S., Miettinen I.T., Savolainen-Kopra C., Pitkänen T. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020:739. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KWR Water Research Institute . 2020. Protocol, SOP Wastewater SARS-CoV-2 RNA Assays. [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020:179. doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugen T.I., Ravelli R.B.G., van Schayck P., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosh S., Kuijpers H.J.H., Schippers D., van de Wetering W., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. eabc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesimple A., Jasim S.Y., Johnson D.J., Hilal N. The role of wastewater treatment plants as tools for SARS-CoV-2 early detection and removal. J. Water Process. Eng. 2020:38. doi: 10.1016/j.jwpe.2020.101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Huang Z., Luo J., Zhang X., Sha S. Primary concentration – the critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: a mini-review. Sci. Total Environ. 2020:747. doi: 10.1016/j.scitotenv.2020.141245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(8):1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhao Y., Liu J., He X., Wang B., Fu S., Yan J., Niu J., Zhou J., Luo B. Effects of temperature variation and humidity on the death of COVID-19 in Wuhan, China. Sci. Total Environ. 2020;724 doi: 10.1016/j.scitotenv.2020.138226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Mao K., Zhang K., Du W., Ali W., Feng X., Zhang H. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr. Opin. Environ. Sci. Health. 2020;17:1–7. doi: 10.1016/j.coesh.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(2020):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari Harmooshi N., Shirbandi K., Rahim F. Environmental concern regarding the effect of humidity and temperature on SARS-COV-2 (COVID-19) survival: fact or fiction. SSRN Electron. J. 2020;27:36027–36036. doi: 10.1007/s11356-020-09733-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem L.D., Morgan B., Donner E., Short M.D. Case Studies in Chemical and Environmental Engineering. Vol. 1. 2020. The COVID-19 pandemic: considerations for the waste and wastewater services sector; p. 100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimer S., Chapman J., Reidy L., Alencar A., Wu Y., Williams S., Pagan L., Gjolah L., MacIntyre J., Triana M., Vance B., Andrews D., Fan Y.-S., Zhou Y., Martinez O., Garcia-Buitrago M., Cray C., Tekin M., McCAuley J., Ruiz P., Pagan P., Lamar W., Alencar M., Bilbao D., Prieto S., Polania M., Suarez M., Lujardo M., Campos G., Morris M., Shukla B., Caban-Martinez A., Kobetz E., Parekh D., Jorda M. A how-to-guide to building a robust SARS-CoV-2 testing program at a university-based health system 2020. medRxiv. 2020;06(03) doi: 10.1101/2020.06.03.20120832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S., Gaddis E., Gray D., Lukasik G., Mull B., Olivas L., Olivieri A., Wu Y. SARS-CoV-2 interlaboratory consortium, reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ. Sci.: Water Res. Technol. 2021;3 doi: 10.1039/D0EW00946F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkin Elmer Commercial COVID-19 Test Kit with Lowest LOD* now Authorized for Symptomatic and Asymptomatic Testing & Pooling. Perkin Elmer. 2021. https://perkinelmer-appliedgenomics.com/home/products/new-coronavirus-2019-ncov-nucleic-acid-detection-kit/
- Perkin Elmer Instructions for PerkinElmer ® New Coronavirus Nucleic Acid Detection Kit, v 8.0. Perkin Elmer. 2021. https://www.fda.gov/media/136410/download
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: Wastewater-based epidemiology for COVID-19 - approaches and challenges for surveillance and prediction. Water Res. 2020;186(Nov 1):116404. doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020:181. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca M.A., Brown R., Solo-Gabriele H.M. Fecal indicator bacteria levels at beaches in the Florida Keys after Hurricane Irma. Mar. Pollut. Bull. 2019;138:266–273. doi: 10.1016/j.marpolbul.2018.09.036. [DOI] [PubMed] [Google Scholar]
- Shibata T., Solo-Gabriele H.M., Fleming L., Elmir S. Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Water Res. 2004;38:3119–3131. doi: 10.1016/j.watres.2004.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman L.M., Gavrilin E., Jorba J., Martin J., Burns C.C., Manor Y., Moran-Gilad J., Sofer D., Hindiyeh M.Y., Gamzu R., Mendelson E., Grotto I. Molecular epidemiology of silent introduction and sustained transmission of wild poliovirus type 1, Israel. Eurosurveillance. 2014;19(7) doi: 10.2807/1560-7917.es2014.19.7.20709. [DOI] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathias V., Koleti A., Vidovic D., Cooper D.J., Jagodnik K.M., Terryn R., Forlin M., Chung C., Torre D., Ayad N., Medvedovic M., Ma'ayan A., Pillai A., Schürer S.C. Sustainable data and metadata management at the BD2K-LINCS Data Coordination and Integration Center. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z., Wang H., Dai Y., Li K., Liu J., Wu W., Yuan C., Yu M., Li P., Yan J. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child. China. Emerg. Infect. Dis. 2020;26(6):1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier J., Darques R., Ait Mouheb N., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health. 2020:10. doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vempati U.D., Chung C., Mader C., Koleti A., Datar N., Erickson S., Muhlich J.L., Berriz G., Benes C.H., Subramanian A., Pillai A., Shamu C.E., Schürer S.C., Vidovic D., Wrobel D. Metadata standard and data exchange specifications to describe, model, and integrate complex and diverse high-throughput screening data from the Library of Integrated Network-based Cellular Signatures (LINCS) J. Biomol. Screen. 2014;19:803–816. doi: 10.1177/1087057114522514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zheng J., Guo L., Yao H., Wang L., Xia X., Zhang W. Fecal viral shedding in COVID-19 patients: clinical significance, viral load dynamics and survival analysis. Virus Res. 2020;289 doi: 10.1016/j.virusres.2020.198147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Mueller M.A., Niemeyer D., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Bruenink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized cases of coronavirus disease 2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alma E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4) doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Wang X., Liu Y., Li G., Qu J. Potential Spreading risks and disinfection challenges of medical wastewater by the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material