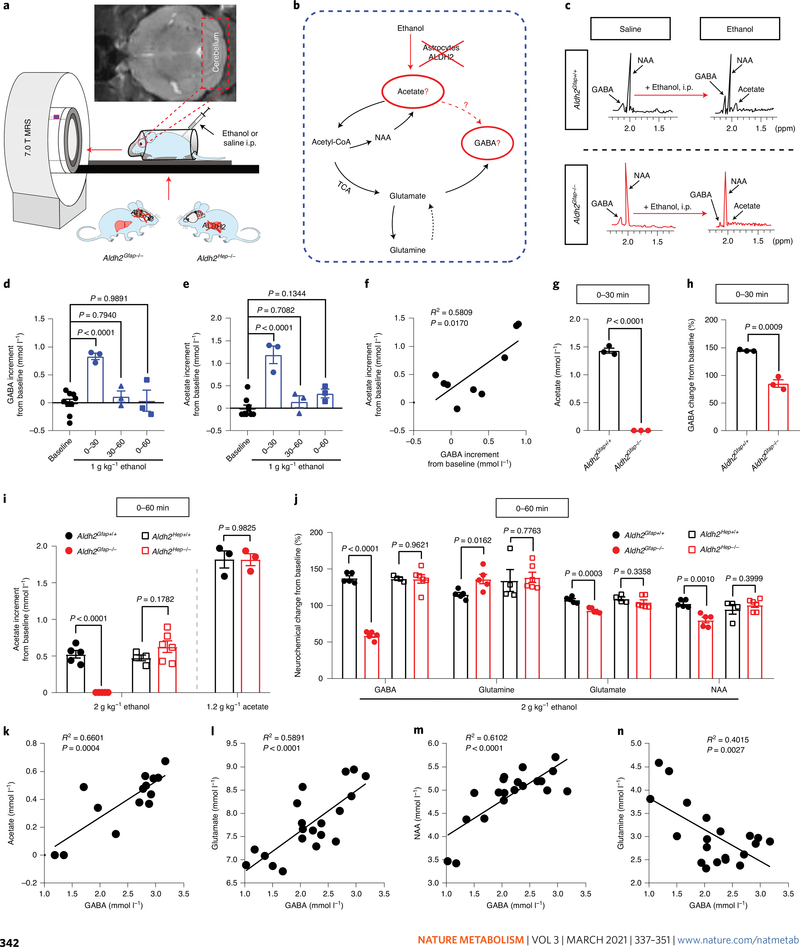
Fig. 3 |. Astrocytic ALDH2 mediates ethanol-induced elevation in cerebellar acetate and GABA levels.
a, Schematic of in vivo quantitative MRS measurement of mouse brain ethanol metabolites. b, Schematic of ethanol–acetate–GABA metabolic pathways. c, Representative spectrometry peaks of cerebellar acetate and GABA before and after i.p. ethanol administration in Aldh2Gfap+/+ mice (upper, black) and Aldh2Gfap−/− mice (lower, red). d, Summary statistics of cerebellar GABA detected by MRS at the different time periods (min) after 1 g kg−1 ethanol administration in Aldh2Gfap+/+ mice (n = 3 mice per group). e, Summary statistics of cerebellar acetate detected by MRS at the different time periods (min) after 1 g kg−1 ethanol administration in Aldh2Gfap+/+ mice (n = 3 mice per group). f, Correlation of cerebellar GABA and acetate after systemic injection of ethanol (P = 0.017 by linear regression). g,h, Summary statistics of GABA and acetate contents by MRS from 0–30 min after systemic injection of 1 g kg−1 ethanol in Aldh2Gfap−/− and Aldh2Gfap+/+ mice. Data were calculated as the percentage change in GABA (before versus after ethanol) in the cerebellum (n = 3 mice per group). i, Summary statistics of cerebellar acetate by MRS from 0–60 min after systemic injection of 2 g kg−1 ethanol or 1.2 g kg−1 acetate in astrocytic ALDH2-deficient mice and in hepatocytic ALDH2-deficient mice (n = 5, 5, 4, 6, 3 and 3 mice, respectively). j, Neurochemical changes after 2 g kg−1 ethanol administration in astrocytic ALDH2-deficient mice (n = 5 mice per group), hepatocytic ALDH2-deficient mice (n = 4 mice in Aldh2Hep+/+ and 6 mice in Aldh2Hep−/−) and their wild-type littermates. Data were calculated as the percentage change in neurochemicals (before versus after ethanol) in the cerebellum. k–n, Correlation analysis of cerebellar GABA with acetate, glutamine, NAA and glutamate detected by MRS (P value was calculated by linear regression). All data are expressed as the mean ± s.e.m. Analysis was performed using an unpaired two-tailed Student’s t-test (g–j) or one-way ANOVA followed by Tukey’s test (d and e).