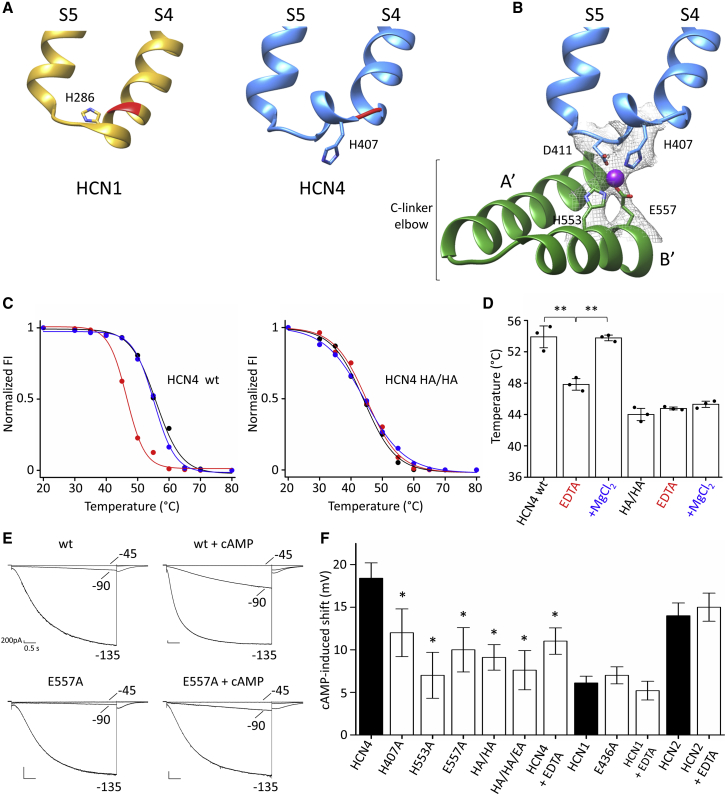
Figure 2.
Structural, biochemical, and functional evidence for the tetrad, connecting the transmembrane and cytosolic domains in HCN4
(A) Comparison of the S4-S5 linker of HCN1 (yellow) and HCN4 (blue). Backbones of I405 in HCN4 and the corresponding I284 in HCN1 are shown in red.
(B) Ribbon representation of S4-S5 linker of one subunit (blue) and underlying C-linker “elbow” (A′–B′ helices) of adjacent subunit (green) of HCN4. Four residues forming the ion coordination site (tetrad) in HCN4 are shown as sticks and labeled (H407, D411, H553, E557). Ion coordinated by the tetrad is represented as a purple sphere.
(C and D) Thermostability assay based on fluorescence-detection size-exclusion chromatography (fSEC-TS) was employed to quantify the change in melting temperature of purified protein in absence or presence of Mg2+ (see STAR Methods). (C) Representative melting curves for holo HCN4 protein purified in LMNG/CHS. Normalized fluorescence intensity (FI) plotted over pre-conditioning temperature (°C). The protein was incubated without (black) or with 10 mM EDTA (red), or, after EDTA removal, with 10 mM MgCl2 (blue). Wild-type protein (HCN4 WT) is shown on the left; double-mutant H407A/H553A (HCN4 HA/HA) is shown on the right. Data points are fitted with sigmoidal dose-response equation (see STAR Methods). (D) Mean denaturation midpoint temperature (Tm) for HCN4 WT in control (53.6°C ± 1.5°C), EDTA (47.8°C ± 0.8°C), + Mg2+ (53.7°C ± 0.4°C); for HCN4 HA/HA: control (44°C ± 0.5°C), EDTA (44.8°C ± 0.1°C), + Mg2+ (45.3°C ± 0.3°C). Values are mean of n = 3 experiments ± SEM. Statistical analysis performed with one-way ANOVA, followed by Fisher’s test (∗∗p < 0.01).
(E) Representative current traces recorded in HEK293 cells expressing WT and mutant E557A HCN4 channels. Patch-clamp recordings were performed in whole-cell configuration, with 30 μM cAMP in pipette solution where indicated. Voltage step protocol was from −30 to −150 mV with −15-mV increments; only currents recorded at −45, −90, and −135 mV are shown. Scale bars: 200 pA × 0.5 s.
(F) cAMP-induced shift (mV) on half activation voltages (V1/2) calculated by fitting a Boltzmann equation to the data (see STAR Methods). Current recordings and activation curves for all WT and mutant channels displayed are shown in Figures S9–S11. V1/2 values are reported in Table S1. Mutations H407A, H553A, and E557A introduced in HCN4, either alone or in combination, reduce the cAMP effect (Figure S9), whereas an equivalent mutation in HCN1 (E436A) does not (see also Figure S10). Addition of 60 mM EDTA to pipette solution of WT channels reduces cAMP response in HCN4 (Figure S9), but not in HCN1 and HCN2 (Figure S11). Values are means of n > 3 experiments ± SEM. Statistical analysis performed with one-way ANOVA, followed by Fisher’s test (∗p < 0.05).