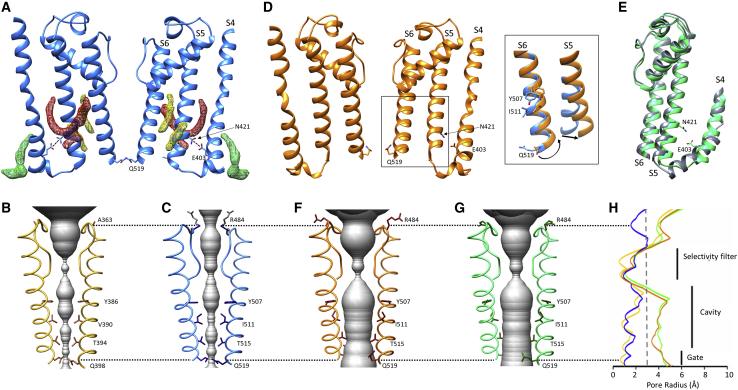
Figure 4.
Transmembrane domain rearrangements in the three HCN4 structures
(A) Ribbon representation of S4, S5, and S6 TM helices in HCN4 holo (blue) in a cross-membrane view. For clarity, only two subunits are shown. Putative lipid densities contacting S4 (green mesh) and S5 (red and yellow mesh) are shown. Residues involved in polar interaction (dotted line) between S4 (E403) and S5 (N421) are shown as sticks and labeled on one subunit. TM helices S4, S5, and S6 are also labeled on one subunit.
(B and C) Ribbon representation of pore of HCN1 holo (yellow, PDB: 5U6P) (B) and HCN4 holo (blue) (C). For ease of view, only two opposite subunits in the assembled tetramer are shown. The pore diameter is shown as a gray surface. Residues facing pore inner cavity and residue of SF facing extracellular side (A363 in HCN1 and R484 in HCN4) are shown as sticks and labeled. The atoms of side chain of R484 whose density is not resolved are colored in gray.
(D) Ribbon representation of S4, S5, and S6 TM helices in HCN4 apo/LC (orange) in a cross-membrane view. For clarity, only two subunits of tetramer are shown. Lipids are absent, and polar interaction between E403 and N421 is lost. Inset: in comparison with HCN4 holo (blue), the apo/LC structure shows a tilting movement of S5 and rotation of S6, as shown by shift in the side chains of Y507, I511, and Q519.
(E) Superimposition of helices S4, S5, and S6 of HCN4 apo/AM (green) and HCN1 in hyperpolarized conformation (gray, PDB: 6UQF). In HCN4 apo/AM, the lipids are absent, and both the polar interaction E403-N421 and hydrophobic interactions between S4 and S5 are lost (see also Figure S10).
(F and G) Ribbon representation of pore of HCN4 apo/LC (orange) (F) and HCN4 apo/AM (green) (G). Labeling as in (B) and (C). Rotation in lower part of S6 relocates side chains of indicated residues away from cavity, widening solvent-accessible pathway (gray) from cytosolic side.
(H), Plot of pore radii, color-coded as in (B), (C), (F), and (G). The dotted line marks the radius of hydrated K+. Corresponding portions of selectivity filter, cavity, and gate are indicated by black vertical bars. Dotted black lines encompassing (B), (C), and (F)–(H) indicate, respectively, R484 in HCN4, the corresponding A363 residue in HCN1, and glutamine residues at the cytosolic gate (Q519 in HCN4, Q398 in HCN1).