Figure 2 .
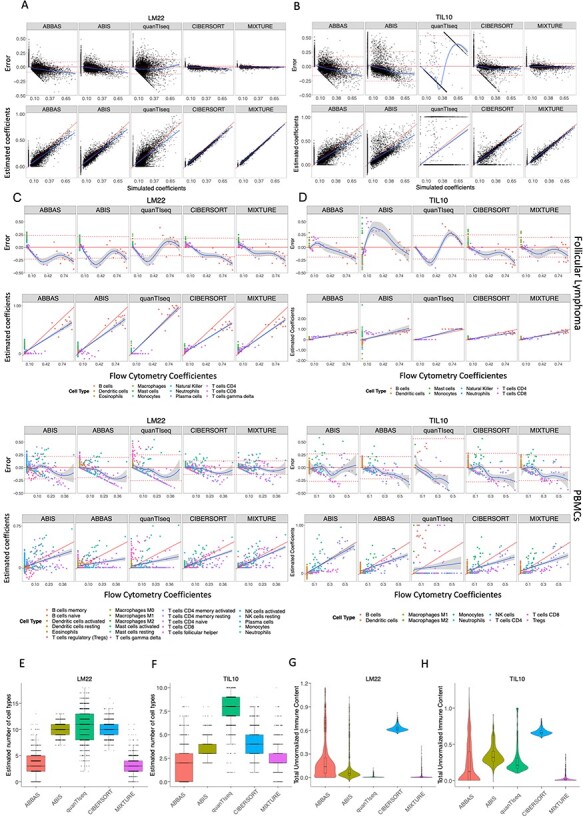
MIXTURE performance on cell type proportion estimation. (A-B) Bland–Altman and correlation plots for Scenario 2 using ABBAS, ABIS, quanTIseq, CIBERSORT and MIXTURE. The upper panels represent the differences between estimated against true simulated values for LM22 and TIL10 signatures, respectively. Horizontal lines indicate the mean ± standard deviation of errors and the loess smooth regression line represents prediction dependent bias. The lower panels show the correlation plots between estimated coefficients and true simulated proportion values. The red line represents the expected identity line, and the blue line is the linear regression line representing prediction depended bias. (C-D) Bland–Altman and correlation plots for Scenario 3 comparing estimated coefficients against true flow cytometry-derived immune content for FL (upper panels) and PBMCs (lower panels) datasets using LM22 and TIL10 signature, respectively by following the validation process from Newman et al. Bland–Altman plots show the difference between estimated coefficients and true coefficients (obtained by flow cytometry). The horizontal continuous and dashed lines represent the overall mean and standard deviation of errors, and the blue continuous line represents the loess smooth regression line to show prediction dependent bias. In correlation plots, the red straight line represents the identity and the blue line represents the regression line between true and estimated proportions. (E-H) For Scenario 4, deconvolution analysis of 1018 pure tumor cell lines from 55 different cancer types obtained from the Cancer Cell Line Encyclopedia (CCLE) was performed with ABBAS, ABIS, CIBERSORT, quanTIseq and MIXTURE methods. In figures E and F, the distribution of the number of total immune cell types estimated by each evaluated method is shown, while figures G and H show the distribution of total un-normalized immune content using LM22 or TIL10 signatures, respectively.
