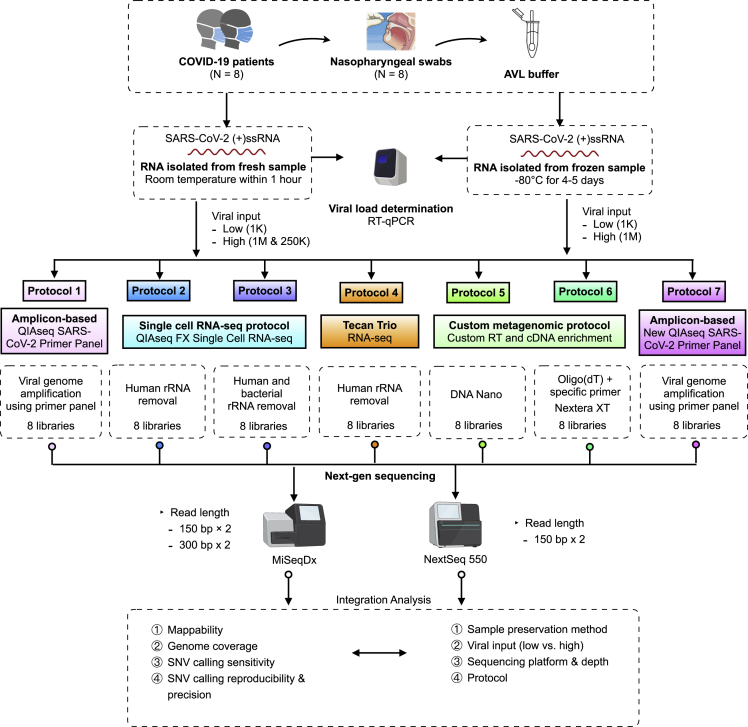
Figure 1.
Schematic overview of the experimental design and workflow
Eight COVID-19 positive patient nasopharyngeal swab samples were used to construct the SARS-CoV-2 WGS libraries using seven protocols. Two different sample storage conditions were compared. For fresh samples, three different viral inputs, i.e., 1000 (1K, low) vs. either 250,000 or 1 million (250K or 1M, high) SARS-CoV-2 viral copies, were used from each same sample, whereas for frozen samples, two different viral inputs, i.e., 1000 (1K, low) vs. 1 million (1M, high) SARS-CoV-2 viral copies from each same sample, were used. P4 used different samples at low input vs. high input due to minimal total RNA amount required. The performances of protocols were benchmarked based on viral input, sequencing platform and depth, mappability, viral genome coverage and coverage uniformity, and sensitivity, reproducibility, as well as precision across seven protocols.