Abstract
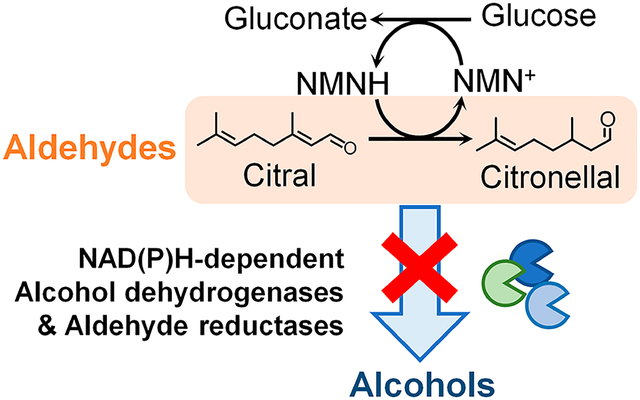
It is challenging to biosynthesize industrially important aldehydes, which are readily consumed by the numerous alcohol dehydrogenases (ADHs) in cells. In this work, we demonstrate that a nicotinamide mononucleotide (NMN+)-dependent redox cofactor cycling system enables aldehyde accumulation in Escherichia coli crude lysates and whole cells. By specifically delivering reducing power to a recombinant enoate reductase, but not to endogenous ADHs, we convert citral to citronellal with minimal byproduct formation (97–100% and 83% product purity in crude lysate- and whole cell-based biotransformation, respectively). We envision the system’s universal application to lowering the noise in biomanufacturing by silencing the host’s metabolic background.
Keywords: nicotinamide mononucleotide, aldehyde, citronellal, noncanonical redox cofactor, Escherichia coli, biomimetic cofactor
Graphical Abstract
INTRODUCTION
Traditionally, biotransformation has been carried out either in vivo with engineered microbial whole cells or in vitro with purified enzymes. Recently, crude lysate-based biotransformation has emerged as an alternative which brings the benefits of both worlds: it contains a cell-like metabolism which includes glycolysis and the TCA cycle that generate energy, cofactors, and key intermediates from inexpensive feedstocks;1–4 it is easier to monitor, manipulate, and optimize because a cell membrane barrier does not exist and operational parameters can be varied well outside what’s tolerable by living cells;5–7 it also eliminates the high cost of purifying proteins for in vitro biotransformation in large scale.1
However, just like in whole cells, crude lysate-based biotransformation faces the challenge of controlling side reactions, which are catalyzed by the numerous endogenous enzymes of the host.1,2 A traditional method to tackle this challenge in whole cells involves identifying and genetically disrupting competing enzymes,8,9 which, albeit effective, can often be time-consuming and labor intensive. Instead of replicating the same approach in a crude lysate-based process for eliminating competing enzymes, here we report an alternative method, which is based on an orthogonal redox cofactor system.10
We chose citronellal as the model product, which is a terpenoid aldehyde with wide usage in the flavor and fragrance industries, as the precursor of nonracemic menthol, and as an insect repellant.11,12 Using enoate reductases (ERs), citronellal can be produced from citral (Figure 1), a low-cost substrate that can be readily obtained in large quantities. However, since both citral and citronellal are aldehydes, which are rapidly reduced to alcohols by a myriad of broad-substrate-range alcohol dehydrogenases (ADHs) in the cell, this conversion is inefficient in microbial hosts in vivo11 and with crude lysates in vitro.13 Previously, Paul and co-workers employed a biomimetic cofactor, 1-benzyl-1,4-dihydronicotinamide, to increase the production of citronellal using poorly purified ER from E. coli.13 This system operated on the basis of the principle that only ER could utilize the biomimetic cofactor as reducing power, while the E. coli endogenous ADHs, which were also present in the crude ER-extract, could not. This method eliminated the cost of extensively purifying ER. However, because this system did not utilize a method of cofactor regeneration, a stoichiometric amount of biomimetic cofactor was required, which could add a formidable cost in large scale. In addition, application of this system in vivo was not examined.
Figure 1.
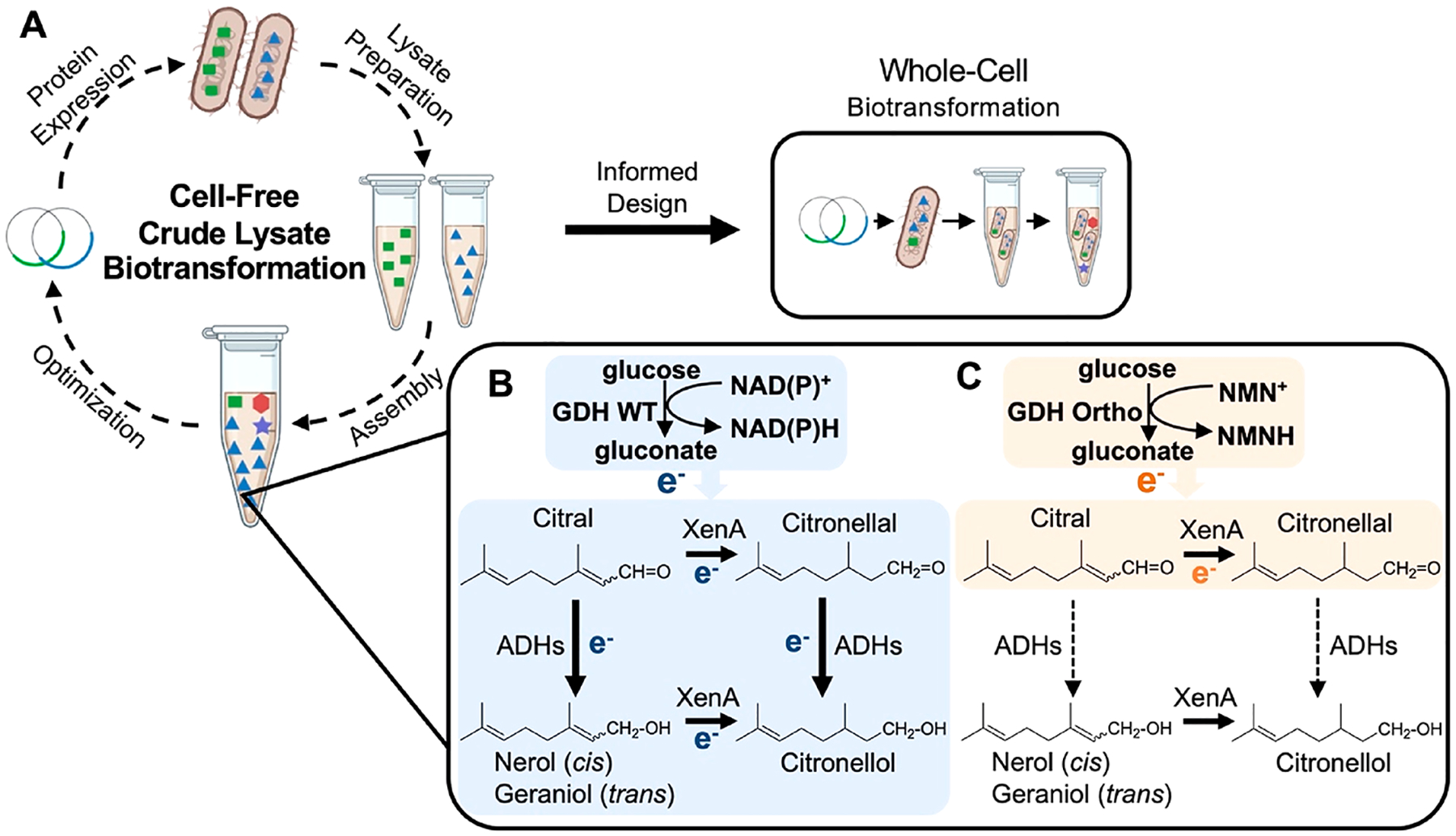
System design and reaction scheme for citronellal production. (A) Crude E. coli lysates are used to form a two-part redox cofactor cycling system which facilitates the conversion of citral to citronellal through the action of Pseudomonas putida XenA. Redox cofactors are continuously regenerated by the glucose dehydrogenase, GDH, from Bacillus subtilis. Information gained from crude lysate-based biotransformation can be used to inform whole cell engineering. (B) When the wild type GDH is supplied, the native cofactors nicotinamide dinucleotide (phosphate), NAD(P)+, are reduced. This supplies the reducing power to both XenA and E. coli ‘s endogenous alcohol dehydrogenases (ADHs), resulting in the formation of alcohols (nerol, geraniol, citronellol) as the final products. (C) When a nicotinamide mononucleotide (NMN+)-specific GDH, GDH Ortho, is introduced, reducing power is only delivered to XenA, which can readily receive NMNH. Endogenous ADH activity is silenced since ADHs cannot receive NMNH. Therefore, citronellal, the target product, will accumulate.
Recently, we demonstrated a nicotinamide mononucleotide (NMN+)-dependent orthogonal redox cofactor system in E. coli, which features an engineered glucose dehydrogenase (GDH Ortho) from Bacillus subtilis, that can efficiently and specifically recycle NMN+ using glucose as an inexpensive substrate.10 In this work, we first demonstrate that the NMN+-based orthogonal redox cofactor system can enable efficient conversion of citral to citronellal in crude lysate-based, cell-free biotransformation without the need to identify or disrupt competing ADHs in E. coli. Next, we systematically optimized enzyme ratios, lysate preparation procedure, cofactor concentration, buffer composition, and pH to achieve citronellal production with 97–100% product purity with nominal levels alcohol byproducts, and ~60% conversion. Finally, the optimized crude lysate system was used to inform the design of whole-cell biotransformation in E. coli (Figure 1A), which produced 33 mg/L citronellal with 83% product purity. In contrast, biotransformation processes relying on the natural cofactors nicotinamide adenine dinucleotide (phosphate) (NAD(P)+) only yielded the alcohol byproducts, namely, citronellol, nerol, and geraniol. We envision this approach may be adapted for different target products and different chassis hosts with relative ease.
RESULTS AND DISCUSSION
Citronellal Production in Crude Lysate-Based Biotransformation.
Using purified ER (Pseudomonas putida XenA) and GDH Ortho in vitro, we have demonstrated citronellal production from citral with NMN+ as the cycling cofactor.10 However, whether this redox cycle is well insulated from the numerous endogenous ADHs in E. coli crude lysates has not been tested. On the basis of previous studies, XenA can accept reducing power in the forms of both NADP(H) and reduced NMN+ (NMNH),10 while endogenous ADHs greatly prefer NAD(P)H over NMNH, likely because of NMNH’s truncated structure which lacks the adenosine moiety “handle” of NAD(P)H.10,14–16 Thus, we hypothesized that when using GDH wild type (WT) to derive reducing power from glucose, both XenA and ADHs will receive electrons because NAD(P)+ are the cycling cofactor (Figure 1B). On the other hand, when GDH Ortho is used with the supplementation of NMN+, only XenA will receive electrons because the stringent cofactor specificity of GDH Ortho allows it to only generate NMNH but not NAD(P)H10 (Figure 1C).
To test this hypothesis, we performed the crude lysate-based biotransformation by paring the XenA module with GDH WT or GDH Ortho module, respectively (Figure 2). GDH WT, GDH Ortho (on plasmids pEK101 or pLZ216, Table S1), and XenA (on plasmid pEK102) were individually overexpressed in E. coli strain MX102 (Table S1). The resulted cells were homogenized to yield three different lysates, which were subsequently mixed-and-matched (Figure 1A). The E. coli host, MX102 (Table S1), contains glycolysis knockouts (Δpgi, Δzwf, ΔgntK) to ensure that glucose is only metabolized by GDH, not the cell’s central metabolism.10 When XenA and GDH WT lysates were mixed at a 1:1 mass ratio, and with the supplementation of no additional cofactor, 1 mM NAD+, or 1 mM NADP+, citronellal only transiently accumulated to a low level (<10 mg/L) before being rapidly consumed (Figure 2A). Notably, the substrate citral also disappeared rapidly (Figure S1). Importantly, when GDH Ortho was used in place of GDH WT, 89 mg/L of citronellal was produced in 4 h with a 39% product purity (the percentage of citronellal in the sum of all products including citronellal, geraniol, nerol, and citronellol) when 1 mM of NMN+ was supplemented (Figure 2A,B). When NMN+ was omitted, citronellal production was not significant, demonstrating that aldehyde accumulation was indeed NMN+ -dependent (Figure 2A). Remarkably, this was achieved without disrupting any aldehyde reductases or ADHs genes.
Figure 2.
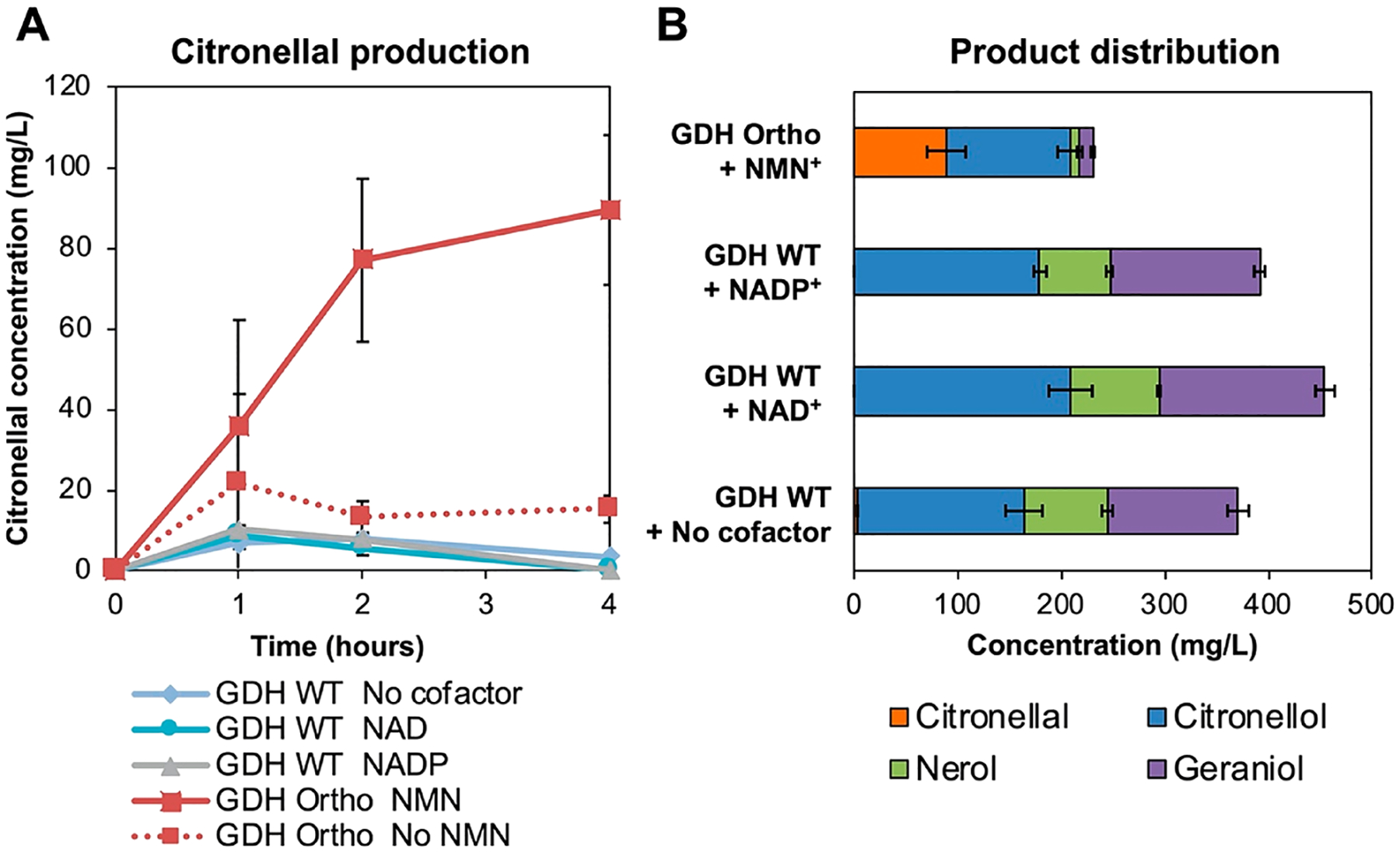
NMN+-dependent orthogonal redox cofactor system enables citronellal accumulation. Crude Escherichia coli lysates enriched with XenA and wild type Bacillus subtilis GDH (GDH WT) or engineered GDH (GDH Ortho) was used to convert citral to citronellal. (A) When GDH WT was used with XenA, citronellal only transiently accumulated to low levels. When GDH Ortho was used with XenA, citronellal was able to accumulate. (B) Product distribution of cycling reactions. The reactions were performed in Buffer A containing 4.5 mg/mL lysates, 1 mM of oxidized cofactor, and 500 mg/L citral. Reactions were incubated at 37 °C. Values are an average of at least three replicates, and the error bars represent one standard deviation.
Optimizing the Crude Lysate-Based Biotransformation.
Although the crude-lysate system was able to accumulate citronellal, key limitations existed (Figure 2B). First, the product purity was suboptimal (~60% of the products were still alcohols, with the major byproduct being citronellol). Second, the conversion was low (89 mg/L citronellal produced from 500 mg/L citral, an ~18% conversion).
We first focused on improving product purity. By running a short-term conversion for 1 h, we used the crude lysate system as a tool to rapidly prototype various ratios of XenA:GDH Ortho lysates,1,17,18 with the total amount of lysates held constant (Figure 3A). We quantified the active enzymes in crude lysates using specific activity assays (Figure S2) and revealed that the 1:1 lysate mass ratio tested in the initial proof-of-concept (Figure 2) contained XenA at a much lower molar concentration than GDH Ortho (8.7 μM XenA versus 23.1 μM GDH Ortho in the reaction system), which led to the hypothesis that XenA might be limiting. Consistent with this hypothesis, increasing the proportion of XenA resulted in increased citronellal production and product purity. The best condition with 7:1 (XenA:GDH Ortho) lysate mass ratio (or 15.1 μM XenA and 5.8 μM GDH Ortho) produced 69 mg/L of citronellal with 16 mg/L citronellol and undetectable levels of geraniol and nerol (Figure 3A), which corresponds to a ~ 78% product purity, a 2-fold improvement compared to the 39% product purity initially obtained using 1:1 ratio (Figure 2B). Citronellal production began to decline when XenA:GDH Ortho was further increased, likely because GDH Ortho became limiting (Figure 3A).
Figure 3.
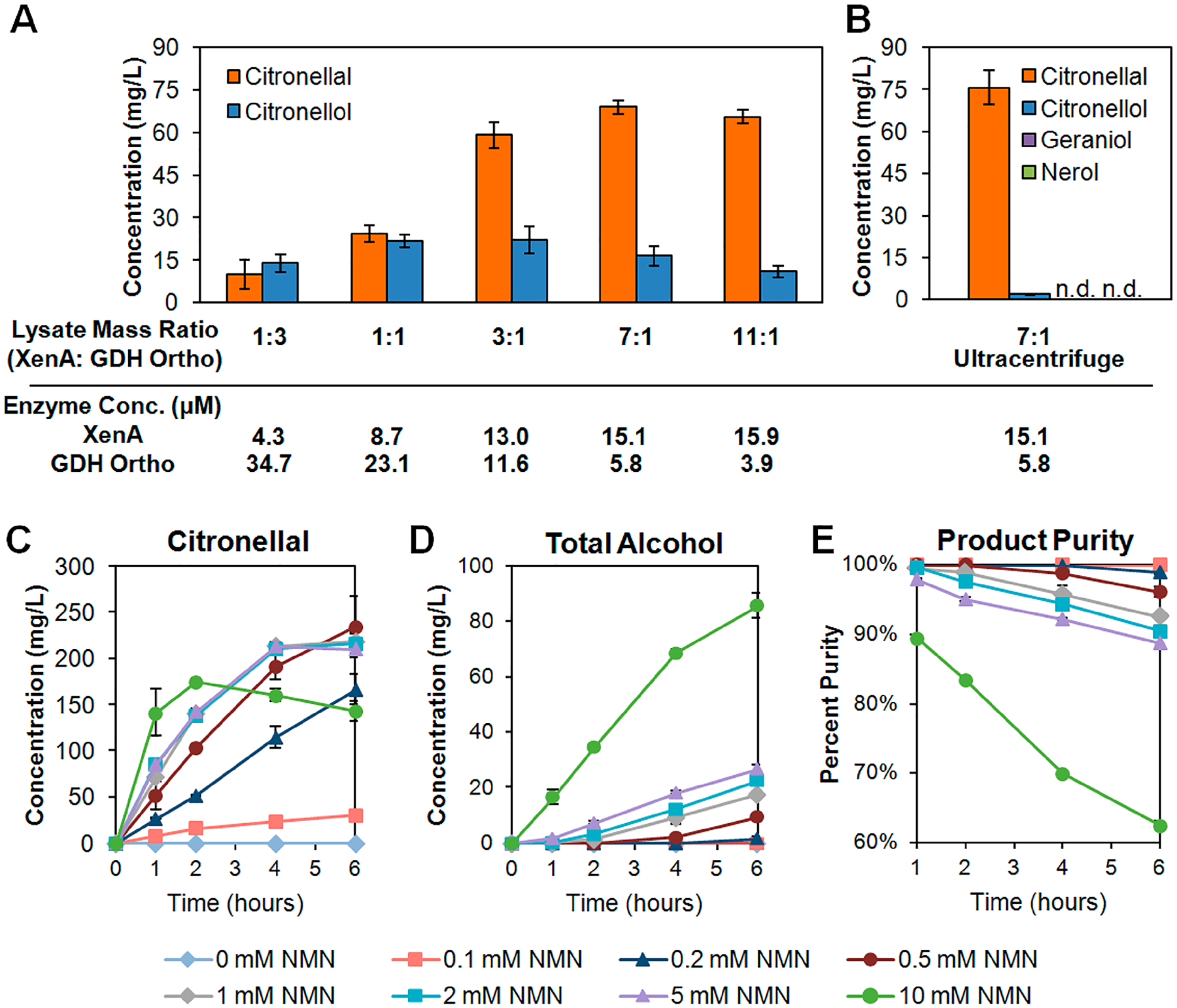
Optimizing crude lysate conditions for citronellal production in Buffer A. (A) Increasing the relative amount of XenA- to GDH Ortho-enriched E. coli crude lysate enabled increased citronellal production and specificity. The molar concentration of each enzyme in the final reaction was calculated for each mass ratio. (B) Further clarifying the lysate with an ultracentrifuge step further improved the performance, yielding a >98% pure production of citronellal. (C–E) Varying NMN+ supplementation in reactions containing ultracentrifuged lysates at a 7:1 XenA:GDH Ortho mass ratio. Product purity was determined as a percentage of citronellal in the total amount of products formed (citronellal, citronellol, nerol, and geraniol). The reactions were performed in Buffer A containing 4.5 mg/mL lysates, 1 mM of NMN+ (unless stated otherwise), and 500 mg/L citral. The reactions were incubated at 37 °C for 1 h unless stated otherwise. Values are an average of at least three replicates, and the error bars represent one standard deviation. n.d., not detected.
When we optimized the lysate preparation to include an ultracentrifugation step, the productivity and purity was further improved (Figure 3B), with 75 mg/L citronellal produced in 1 h with a 98% product purity. Alcohol byproduct formation was minimized, with <2 mg/L citronellol and undetectable levels of geraniol and nerol produced (Figure 3B). However, this high purity could not be maintained as the conversion goes beyond 1 h (Figure 3C–E, gray line with 1 mM NMN+), which motivated further optimization.
Next, we examined the effect of NMN+ concentration (Figure 3C–E). Increasing NMN+ supplementation from 1 mM to 2, 5, or 10 mM did not improve final citronellal titer or product purity because significant amount of alcohols (citronellol, nerol, and geraniol) were produced (Figure 3C–E, Figure S3). Further investigation is needed to identify the cause of increased alcohols, but one possible explanation could be that ADHs may be able to utilize NMNH only at a very high concentration. Although this NMNH-dependent ADH activity is much slower compared to the NMNH-dependent ER activity, at later time points, the former became significant. On the other hand, decreasing NMN+ supplementation to 0.1, 0.2, and 0.5 mM enhanced the product purity (Figure 3E). Notably, a 0.1 mM NMN+ supplementation yielded a 100% product purity with nondetectable levels of alcohol byproducts throughout the entire conversion period. However, the productivity was slow, which led to a low final titer of ~30 mg/L citronellal before the reaction plateaued at 6 h (Figure 3C–E).
We next focused on improving the final citronellal titer and percent conversion (amount of citronellal produced over that of citral supplied), without sacrificing product purity. First, by switching the buffer system in the reaction from Buffer A to phosphate buffer, we increased the citronellal titer by roughly 4-fold, reaching ~114 mg/L, while still maintaining 100% product purity (Figure 4A). Second, we optimized the phosphate buffer conditions by varying both NMN+ concentrations and pH simultaneously (Figure 4B–D). In light of the results mentioned above, NMN+ titration focused on the lower concentration range.
Figure 4.
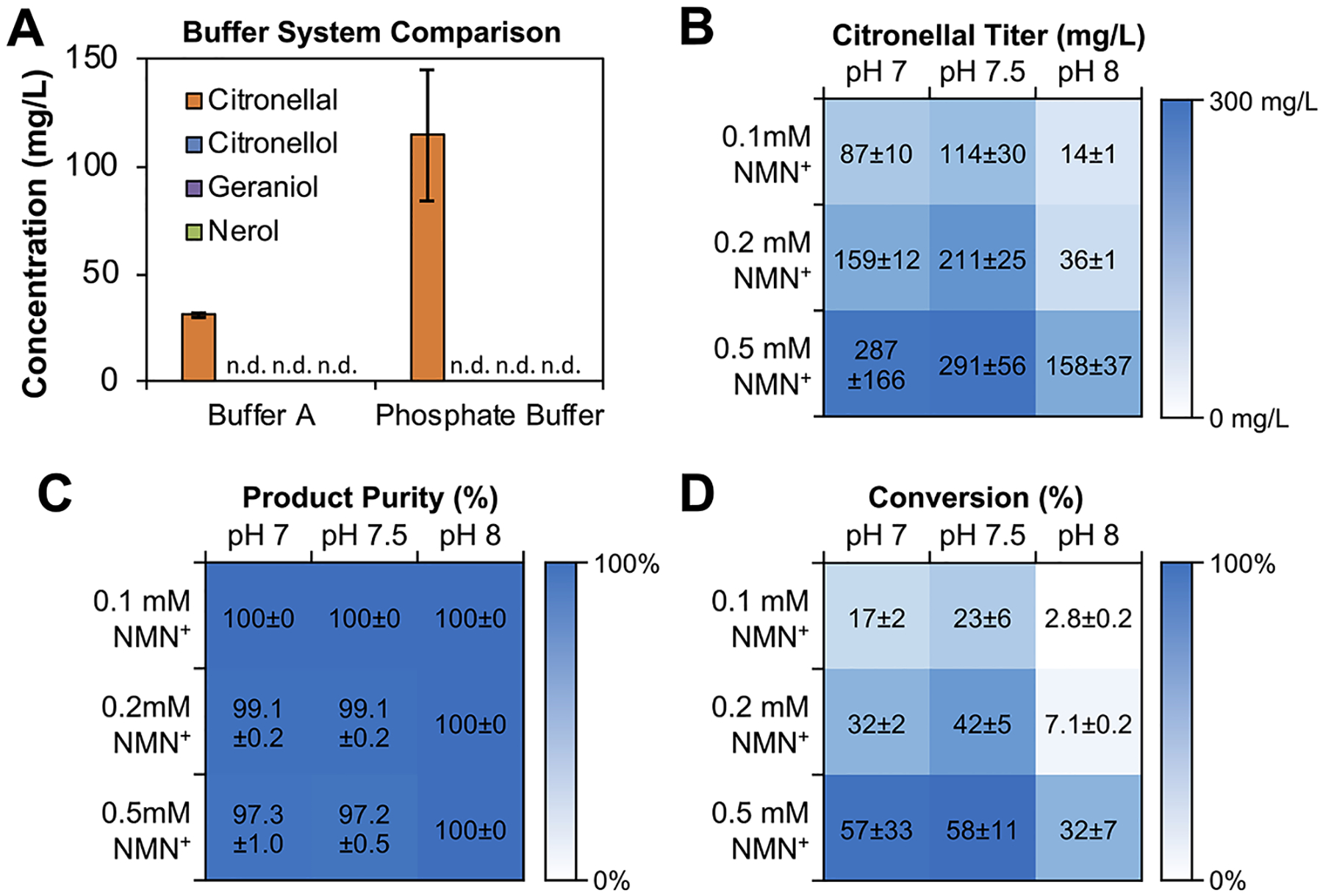
Crude lysate-based biotransformation in potassium phosphate buffer. (A) Performing the crude lysate-based biotransformations in phosphate buffer at pH 7.5 increased citronellal production over Buffer A without producing alcohol byproducts after 6 h. Reactions were supplemented with 0.1 mM NMN+. (B–D) Optimizing pH and NMN+ supplementation for citronellal production, product purity, and conversion. Product purity was determined as a percentage of citronellal in the total amount of products formed (citronellal, citronellol, nerol, and geraniol). Conversion was determined as a percentage of citronellal produced from the 500 mg/L of citral supplied. Phosphate buffered reactions contained 200 mM potassium phosphate, 200 mM sodium chloride, 200 mM d-glucose, 0.1–0.5 mM NMN+, and 500 mg/L citral. All reactions contained 4.5 mg/mL ultracentrifuged lysates at a 7:1 XenA:GDH Ortho mass ratio and were incubated at 37 °C for 6 h. Values are an average of at least three replicates, and the error bars represent one standard deviation.
We identified the best condition at 0.5 mM NMN+ and pH 7.5 in phosphate buffer (with ultracentrifuged lysates, and an enzyme loading of 15.1 μM XenA and 5.8 μM GDH Ortho, as previously mentioned). In this condition, ~292 mg/L citronellal was produced, resulting in ~60% conversion. Product purity was ~97%, with <9 mg/L citronellol and undetectable levels of geraniol and nerol formed (Figure S4). In our previous work using purified GDH and XenA at a similar enzyme loading, we achieved ~75% conversion from citral to citronellal using NMN+.10 Alcohol byproducts were not produced in that process because ADHs were removed during protein purification. Using crude lysates here, we were able to recapitulate the high purity and a comparable conversion, while bypassing the need to eliminate any ADHs or purify any proteins.
Citronellal Production in E. coli Whole Cells.
We next examined the ability of the orthogonal cofactor system to produce citronellal in vivo in E. coli whole cells. Previously, crude lysate-based biotransformation has been shown as a powerful tool to prototype and inform pathway design in whole-cells.1,3,19 Here, we demonstrated the benefit of maintaining a high XenA:GDH Ortho ratio is translatable from in vitro to in vivo. To modulate the XenA:GDH ratio, XenA and GDH were expressed in strain MX102 on two, multicopy plasmids with different copy numbers and promoters (PBAD RSF ori versus PLlacO1 ColE1 ori). The relative expression level of the two plasmids was determined by measuring GDH Ortho’s NMN+ reducing activity in crude lysate derived from the whole cells (Figure 5A). When GDH Ortho was expressed on the PLlacO1 ColE1 ori vector (pLZ216), the activity was ~7-fold higher than on the PBAD RSF ori vector (pSM106) (Figure 5A). Therefore, the PBAD RSF ori or PLlacO1 ColE1 ori vectors were designated as the low and high expression vectors, respectively, for in vivo biotransformation.
Figure 5.
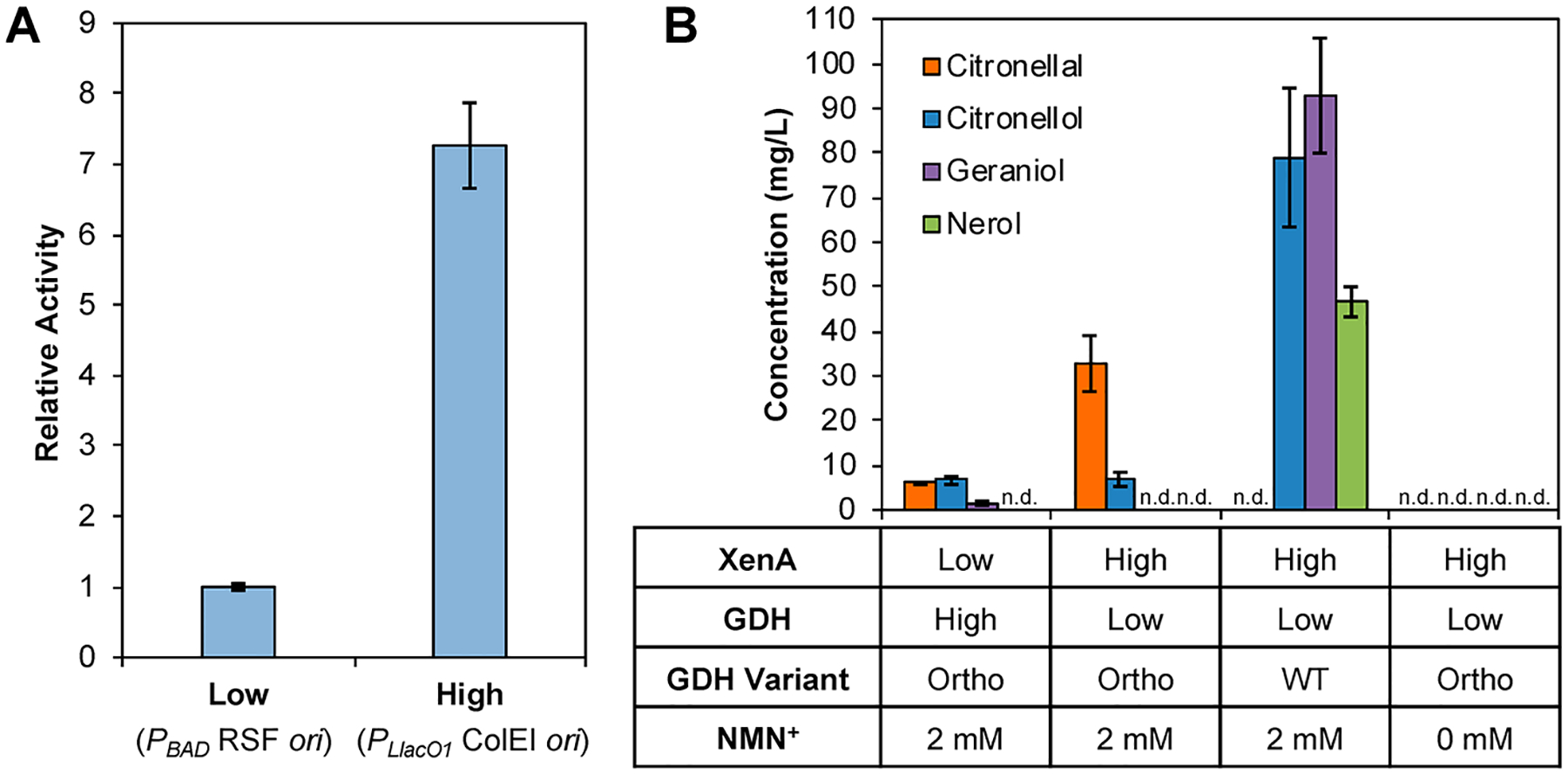
Orthogonal cofactor system enables specific aldehyde accumulation in E. coli whole cells. (A) Relative expression levels of PBAD RSF ori and PLlacO1 ColE1 ori vectors as measured by the levels of GDH Ortho activity in crude lysate derived from the coexpressed whole cells. The results indicate that PLlacO1 ColE1 ori and PBAD RSF ori vectors can be treated as a high and low expression vectors, respectively. (B) With GDH Ortho, citronellal accumulation was achieved. High XenA expression relative to GDH Ortho resulted in higher citronellal productivity and product purity. With GDH WT, only alcohol byproducts were produced. When NMN+ was not supplemented to cells expressing GDH Ortho, no product accumulation was observed. Whole-cell biotransformation was performed with resting E. coli cells at an OD600 of 10 in 100 mM potassium phosphate at pH 7.5, 200 mM d-glucose, 2 mM NMN+, and 500 mg/L citral, at 30 °C for 3 h while shaking at 250 r.p.m. Values are an average of at least three replicates, and the error bars represent one standard deviation. n.d., not detected.
When resting E. coli whole cells were supplied with high XenA expression (on plasmid pEK102, Table S1), with low GDH Ortho expression (on plasmid pSM106), and a glucose facilitator, Zymomonas mobilis Glf (on plasmid pSM109) which transports glucose into the cell, 33 mg/L of citronellal was produced with a product purity of 83% (Figure 5B). Conversely, pairing high GDH Ortho expression with low XenA expression (on plasmids pLZ216 and pLZ217, respectively) caused a significant decrease in citronellal production (6 mg/L), and a poor product purity of 42%. In both conditions, the major byproduct citronellol was formed at 7 mg/L. When GDH Ortho was not supplied with NMN+, citronellal production was not observed (Figure 5B). Expectedly, when GDH WT was supplied (on plasmid pSM107), endogenous ADH activity completely consumed citral and citronellal, resulting in the production of only geraniol, nerol, and citronellol (Figure 5B). Extending the reaction time to 24 h did not provide a higher conversion or product purity than that achieved after 3 h (Figure S5). Ongoing work in our lab is focused on further optimizing the whole-cell system.
CONCLUSIONS
This work demonstrates the use of an NMN+-dependent redox cofactor system as a facile method to develop biocatalysts for aldehyde production. As a proof-of-concept, we achieved effective conversion of citral to citronellal (both are aldehydes) by silencing the highly active ADHs in both E. coli crude lysates and whole cells. Notably, we decreased the level of alcohol byproducts in crude lysate-based biotransformation from 60% to essentially zero by only optimizing the components in the NMN+ redox loop and without relying on information about the host’s metabolic background. The ease in optimization highlights the advantage of an insulated, orthogonal metabolic system, which may be particularly beneficial in engineering nonmodel organisms without well understood metabolisms. As more enzymes are engineered to efficiently utilize NMN(H), the methods established here may be readily adapted to produce various chemicals.
The orthogonal redox cofactor system is highly complementary to the commonly used host engineering methods. For example, while the orthogonal redox cofactor system bypassed the laborious process of knocking out the numerous, highly promiscuous ADHs and aldehyde reductases,20,21 efforts to genetically eliminate the fewer, more substrate-specific, and more easily identifiable competing enzymes that consume citral and citronellal (namely, aldehyde dehydrogenases and citral lyase) may further improve biotransformation performance.11,22
Supplementary Material
ACKNOWLEDGMENTS
H.L. acknowledges support from the University of California, Irvine, the National Science Foundation (NSF) (award no. 1847705), and the National Institutes of Health (NIH) (award no. DP2 GM137427). W.B.B. acknowledges support from Graduate Assistance in Areas of National Need fellowship funded by the U.S. Department of Education. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the NSF.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscatal.0c03070.
Experimental methods, plasmids, and strains used in this study (Table S1), accession numbers for proteins used in this study (Table S2), citral consumption in crude lysate-based biotransformation (Figure S1), standard curves used to determine recombinant protein concentration in crude lysates (Figure S2), time-course of citral biotransformation and optimization in Buffer A (Figure S3), Time-course of citral biotransformation and optimization in phosphate buffer (Figure S4), resting E. coli whole-cell biotransformation 24 h time point, and references for the Supporting Information (PDF)
The authors declare no competing financial interest.
Contributor Information
Kelly N. Richardson, Department of Chemical and Biomolecular Engineering, University of California Irvine, Irvine, California 92697-2700, United States.
William B. Black, Department of Chemical and Biomolecular Engineering, University of California Irvine, Irvine, California 92697-2700, United States.
Han Li, Department of Chemical and Biomolecular Engineering, University of California Irvine, Irvine, California 92697-2700, United States;.
REFERENCES
- (1).Karim AS; Dudley QM; Juminaga A; Yuan Y; Crowe SA; Heggestad JT; Garg S; Abdalla T; Grubbe WS; Rasor BJ; Coar DN; Torculas M; Krein M; Liew F; Quattlebaum A; Jensen RO; Stuart JA; Simpson SD; Köpke M; Jewett MC In vitro Prototyping and Rapid Optimization of Biosynthetic Enzymes for Cell Design. Nat. Chem. Biol 2020, 16, 912–919. [DOI] [PubMed] [Google Scholar]
- (2).Dudley QM; Nash CJ; Jewett MC Cell-free Biosynthesis of Limonene Using Enzyme-Enriched Escherichia coli Lysates. Synth. Biol 2019, 4, No. ysz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Karim AS; Jewett MC A Cell-Free Framework for Rapid Biosynthetic Pathway Prototyping and Enzyme Discovery. Metab. Eng 2016, 36, 116–126. [DOI] [PubMed] [Google Scholar]
- (4).Kay JE; Jewett MC Lysate of Engineered Escherichia coli Supports High-Level Conversion of Glucose to 2, 3-butanediol. Metab. Eng 2015, 32, 133–142. [DOI] [PubMed] [Google Scholar]
- (5).Kay JE; Jewett MC A Cell-Free System for Production of 2,3-Butanediol is Robust to Growth-Toxic Compounds. Metab. Eng. Commun 2020, 10, No. e00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Honda K; Kimura K; Ninh PH; Taniguchi H; Okano K; Ohtake H In vitro Bioconversion of Chitin to Pyruvate with Thermophilic Enzymes. J. Biosci. Bioeng 2017, 124, 296–301. [DOI] [PubMed] [Google Scholar]
- (7).Wang W; Liu M; You C; Li Z; Zhang YHP ATP-Free Biosynthesis of a High-Energy Phosphate Metabolite Fructose 1,6-diphosphate by in vitro Metabolic Engineering. Metab. Eng 2017, 42, 168–174. [DOI] [PubMed] [Google Scholar]
- (8).Kunjapur AM; Tarasova Y; Prather KLJ Synthesis and Accumulation of Aromatic Aldehydes in an Engineered Strain of Escherichia coli. J. Am. Chem. Soc 2014, 136, 11644–11654. [DOI] [PubMed] [Google Scholar]
- (9).Rodriguez GM; Atsumi S Toward Aldehyde and Alkane Production by Removing Aldehyde Reductase Activity in Escherichia coli. Metab. Eng 2014, 25, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Black WB; Zhang L; Mak WS; Maxel S; Cui Y; King E; Fong B; Sanchez Martinez A; Siegel JB; Li H Engineering a Nicotinamide Mononucleotide Redox Cofactor System for Biocatalysis. Nat. Chem. Biol 2020, 16, 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hall M; Hauer B; Stuermer R; Kroutil W; Faber K Asymmetric Whole-Cell Bioreduction of an α,β-Unsaturated Aldehyde (Citral): Competing prim-Alcohol Dehydrogenase and C-C Lyase Activities. Tetrahedron: Asymmetry 2006, 17, 3058–3062. [Google Scholar]
- (12).Mahalwal VS; Ali M Volatile Constituents of Cymbopogon nardus (Linn.) Rendle. Flavour Fragrance J 2003, 18, 73–76. [Google Scholar]
- (13).Paul CE; Gargiulo S; Opperman DJ; Lavandera I; Gotor-Fernández V; Gotor V; Taglieber A; Arends IWCE; Hollmann F Mimicking Nature: Synthetic Nicotinamide Cofactors for C = C Bioreduction Using Enoate Reductases. Org. Lett 2013, 15, 180–183. [DOI] [PubMed] [Google Scholar]
- (14).Sicsic S; Durand P; Langrene S; le Goffic F A New Approach of Using Cofactor Dependent Enzymes: Example of Alcohol Dehydrogenase. FEBS Lett 1984, 176, 321–324. [DOI] [PubMed] [Google Scholar]
- (15).Campbell E; Meredith M; Minteer SD; Banta S Enzymatic Biofuel Cells Utilizing a Biomimetic Cofactor. Chem. Commun 2012, 48, 1898–1900. [DOI] [PubMed] [Google Scholar]
- (16).Josa-Culleré L; Lahdenperä ASK; Ribaucourt A; Höfler GT; Gargiulo S; Liu Y; Xu J; Cassidy J; Paradisi F; Opperman DJ; Hollmann F; Paul CE Synthetic Biomimietic Coenzymes and Alcohol Dehydrogenases for Asymmetric Catalysis. Catalysts 2019, 9, 207. [Google Scholar]
- (17).Dudley QM; Anderson KC; Jewett MC Cell-Free Mixing of Escherichia coli Crude Extracts to Prototype and Rationally Engineer High-Titer Mevalonate Synthesis. ACS Synth. Biol 2016, 5, 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Liu Z; Zhang Y; Jia X; Hu M; Deng Z; Xu Y; Liu T In vitro Reconstitution and Optimization of the Entire Pathway to Convert Glucose to Fatty Acid. ACS Synth. Biol 2017, 6, 701–709. [DOI] [PubMed] [Google Scholar]
- (19).Kelwick R; Ricci L; Chee SM; Bell D; Webb AJ; Freemont PS Cell-Free Prototyping Strategies for Enhancing the Sustainable Production of Polyhydroxyalkanoates Bioplastics. Synth. Biol 2018, 3, No. ysy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhou YJ; Buijs NA; Zhu Z; Gomez DO; Boonsombuti A; Siewers V; Nielsen J Harnessing Yeast Peroxisomes for Biosynthesis of Fatty-Acid-Derived Biofuels and Chemicals with Relieved Side-Pathway Competition. J. Am. Chem. Soc 2016, 138, 15368–15377. [DOI] [PubMed] [Google Scholar]
- (21).Lau YH; Giessen TW; Altenburg WJ; Silver PA Prokaryotic Nanocompartments Form Synthetic Organelles in a Eukaryote. Nat. Commun 2018, 9, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Esmaeili A; Rohany S; Safaiyan S Biotransformation of Citral by Free and Immobilized Saccharomyces cerevisiae. Chem. Nat. Compd 2012, 48, 322–324. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


