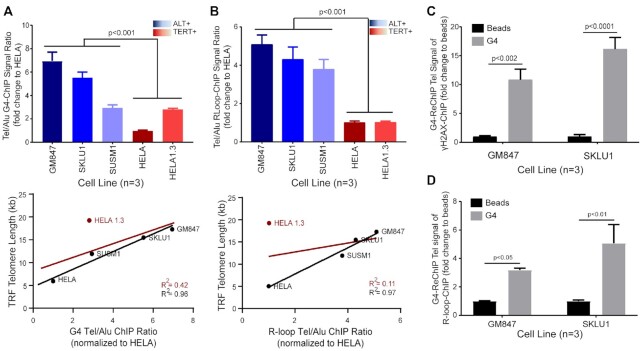
Figure 2.
Levels of telomeric G4s and R-loops are higher in ALT+ cells and are broadly associated with telomere length and coupled with DNA damage signals. (A andB) Top; (A) G4-ChIP assay or (B) R-loop-ChIP assay in ALT+ cell lines (blue shades) versus TERT+ cell lines (red shades). Data shown are the means from three biological repeat experiments. T-test (two-tailed) was performed for the combined ALT+ versus TERT+ datasets. Bottom; linear regression plot of Tel/Alu ChIP signal ratio against telomere length (as measured by telomere restriction fragment (TRF) analysis). Line of best fit is plotted along with R2 values with HELA1.3 (red) or without HELA1.3 (black). (C) ChIP-Re-ChIP (γH2AX-G4) and quantified for telomeric DNA (Tel DNA) in ALT+ cells, GM847 and SKLU1. The ReChIP signal for each sample was then normalized to its respective beads only control. Two-way ANOVA with Sidak multiple correction was performed. Data shown are the means from three biological repeat experiments. Error bars represent the SEM. (D) ChIP-ReChIP (first ChIP for R-loop followed by second ChIP for G4) and quantified for Tel DNA in ALT+ cells, GM847 and SKLU1. ReChIP signal for each sample was then normalized to its respective beads only control. Two-way ANOVA with Sidak multiple correction was performed. Data shown are the means from three biological repeat experiments. Error bars represent the SEM.