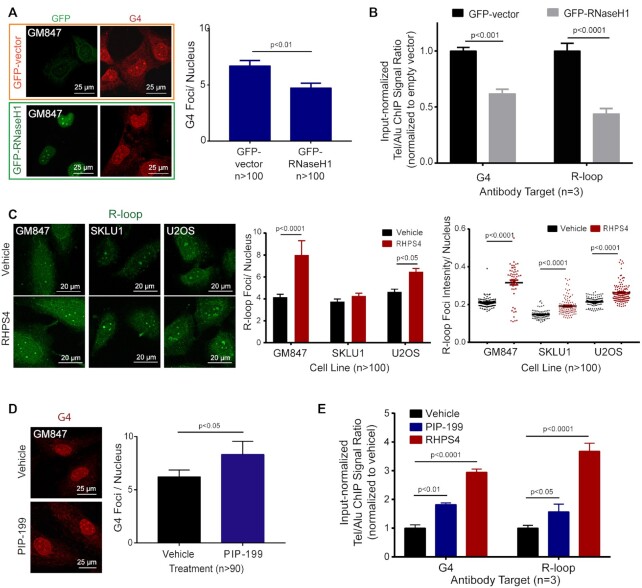
Figure 3.
G4s and R-loops are interdependent. (A) ICC staining for GFP (green) and G4 (red) in GM847 transfected with GFP-vector control (within orange frame) or GFP-RNaseH1 (within green frame). Right, Quantification of GFP+ cells for mean BG4 foci count per nucleus. T-test (two-tailed) was performed. (B) G4-ChIP or R-loop-ChIP results from GM847 transfected with either GFP-vector or GFP-RNaseH1. Tel/Alu ChIP signals were then normalized against GFP-vector control. Data shown are the mean from three biological repeat experiments. Two-way ANOVA with Sidak multiple comparisons was performed. Error bars represent the SEM. (C) Left, R-loop ICC staining (green) in ALT+ cell lines (GM847, SKLU1, U2OS) treated with vehicle or G4 ligand, RHPS4 (1, 1.5 and 5 μM, respectively). Mean R-loop foci count (center) and intensity (right) per nucleus were quantified. About 100 cells per treatment were quantified. Two-way ANOVA with Sidak-corrected multiple comparisons was performed against the untreated controls. (D) G4 ICC staining (red) in GM847 with vehicle or R-loop inducer, PIP-199 0.75 μM. A total of 50 cells per treatment were quantified. T-test (two-tailed) was performed. (E) G4-ChIP or R-loop-ChIP of GM847 treated with vehicle, 0.75 μM PIP-199 or 1 μM RHPS4. ChIP signal was first normalized to input then the Tel/Alu ratio calculated for each condition was then normalized against that of the vehicle control. Data shown are the mean from three biological repeat experiments. Two-way ANOVA with Sidak-corrected multiple comparisons was performed against the vehicle control. Error bars represent the SEM.