Fig. 1. Transmembrane proteome profiling of the region-resolved mouse brain with DIA MS analysis.
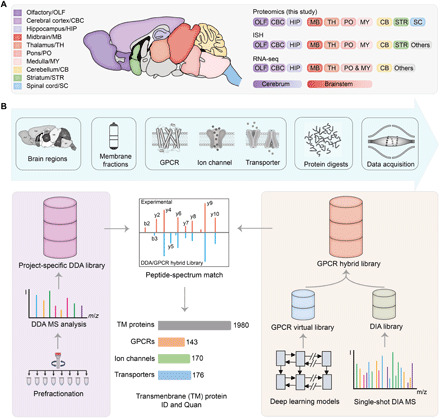
(A) A summary of the brain regions of the resting-state mice examined in this study (left) and the overlapping regions analyzed using ISH by Allen Brain Atlas or RNA-seq by Mulder and colleagues (6) (right). (B) Overall workflow of mouse brain DIA MS analysis and data mining. For each brain region, membrane proteins [represented by three transmembrane protein families this study focuses on] were isolated, extracted, and digested before MS analysis (top). A project-specific DDA library was built from DDA MS analysis of prefractionated multiregional brain tissues (left route). Meanwhile, a GPCR family–targeted hybrid library was built by merging an initial DIA library derived from the DIA MS data and a GPCR virtual library predicted from 524 mouse genome–encoded GPCR sequences using deep learning models (right route). The DIA MS data were searched with the two libraries to yield transmembrane protein ID and quantification results.
