Fig. 2. Deep proteome coverage and reproducible quantification of GPCR, ion channel, and transporter family members in the mouse brain.
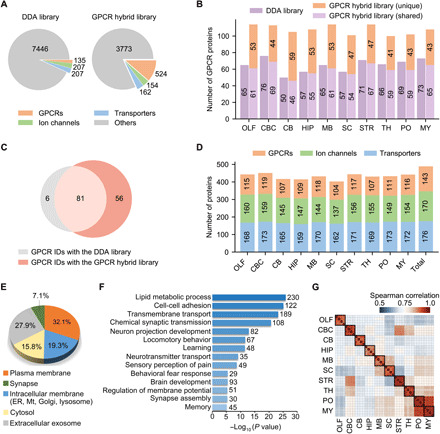
(A) Number of protein IDs in the DDA library and the GPCR hybrid library. The full complement of 524 genome-encoded GPCRs are included in the latter. (B) Number of GPCR IDs in each brain region yielded with the DDA library (purple) or the GPCR hybrid library after data filtering (light purple, shared IDs between the two libraries; orange, unique IDs only yielded with the hybrid library). OLF, olfactory bulb; CBC, cerebral cortex; CB, cerebellum; HIP, hippocampus; MB, midbrain; SC, spinal cord; STR, striatum; TH, thalamus; PO, pons; MY, medulla. (C) Comparison of GPCR IDs from 10 brain regions yielded with the two libraries. (D) Number of protein IDs for three families in each region and in total. GPCR IDs are concatenated from two libraries, and ion channel and transporter IDs were detected with the DDA library. (E) Subcellular localization of all protein IDs according to gene ontology cellular component classification. ER, endoplasmic reticulum; Mt, mitochondria. (F) Many enriched biological processes (P < 10−5) in all protein IDs are related to neuronal cell activity or brain functions. (G) Spearman correlation of protein quantification between replicates of each region.
