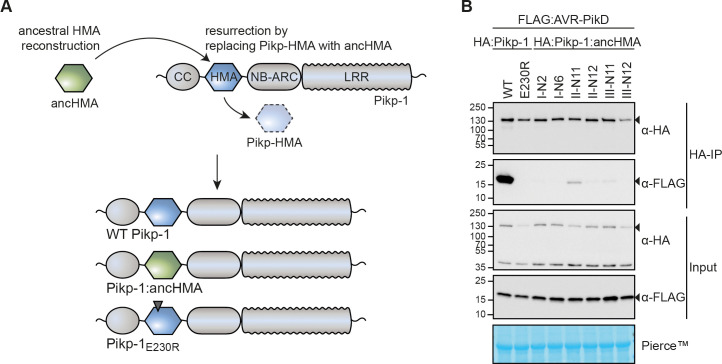
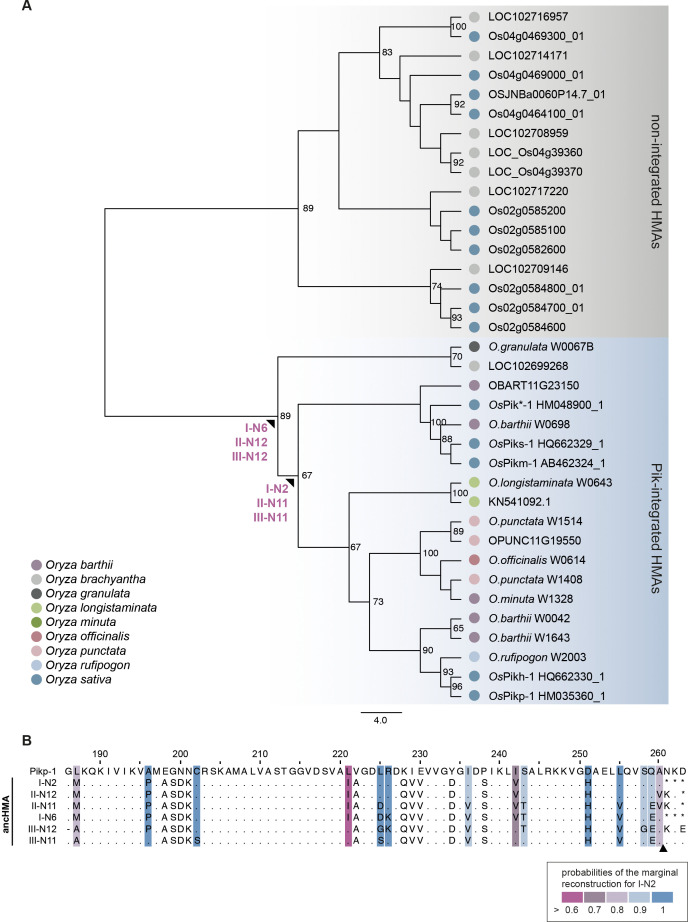
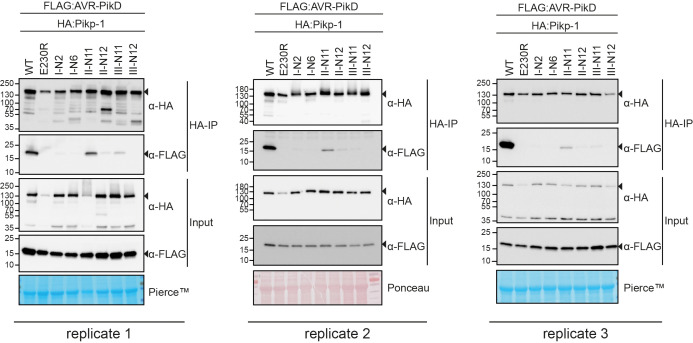
Figure 3. The integrated heavy metal-associated (HMA) domain of Pikp-1 exhibits stronger association with the AVR-PikD effector than its predicted ancestral state.
(A) Overview of the strategy for resurrection of the ancestral HMA (ancHMA) domain. Following ancestral sequence reconstruction, the gene sequences were synthesised and incorporated into Pikp-1 by replacing the present-day Pikp-HMA domain (blue) with the ancHMA equivalent (green). (B) Co-immunoprecipitation experiment between AVR-PikD (N-terminally tagged with FLAG) and Pikp-1 (N-terminally tagged with HA) carrying ancestral sequences of the HMA. Wild-type (WT) HA:Pikp-1 and HA:Pikp-1E230R were used as a positive and negative control, respectively. Immunoprecipitates (HA-IP) obtained with anti-HA probe and total protein extracts (Input) were immunoblotted with appropriate antisera (listed on the right). Rubisco loading control was performed using Pierce staining solution. Arrowheads indicate expected band sizes. Results from three independent replicates of this experiment are shown in Figure 3—figure supplement 3.