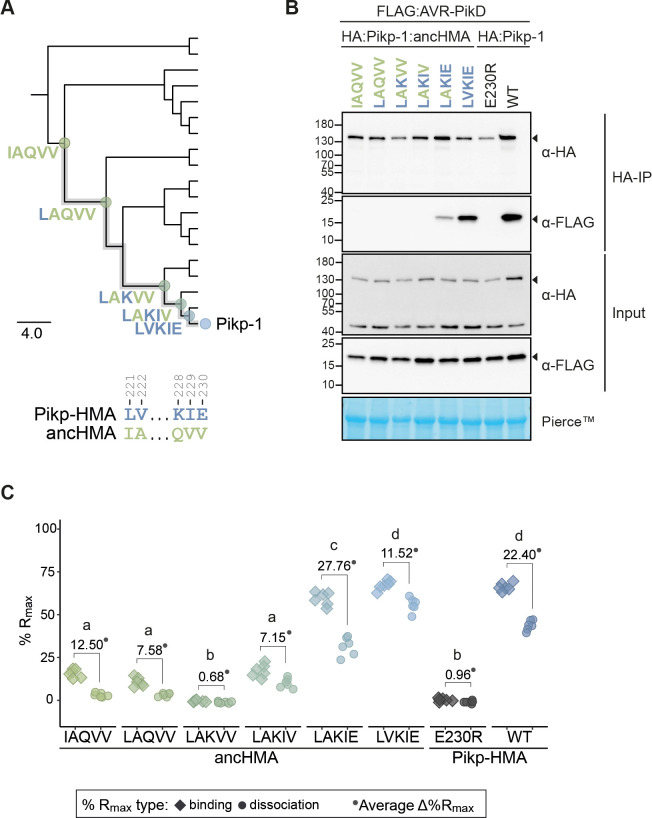
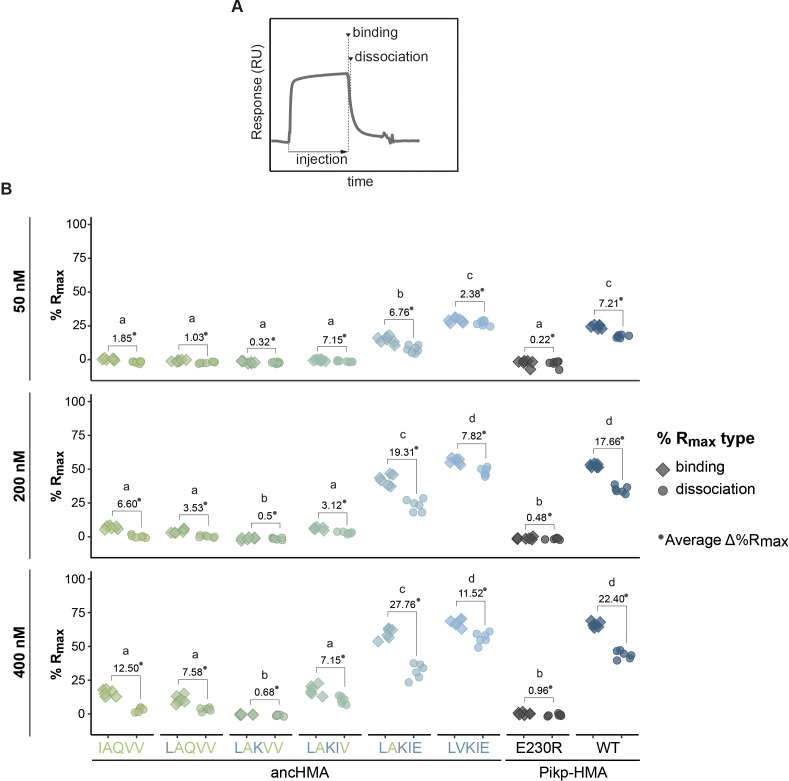
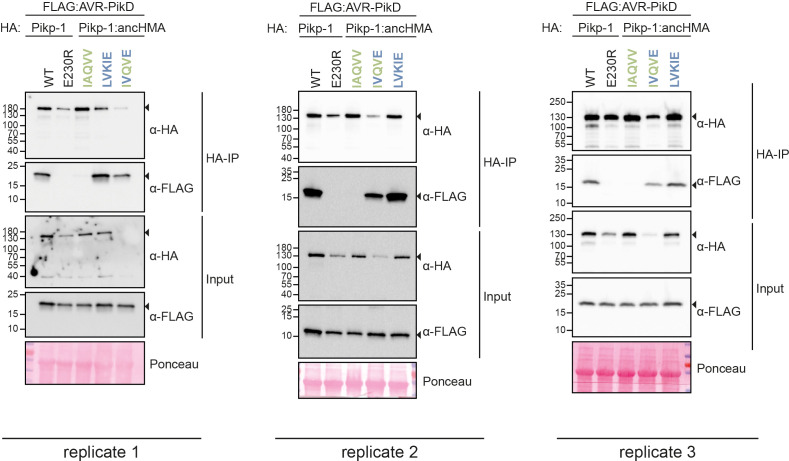
Figure 5. The AV-VE substitutions within the IAQVV/LVKIE region of ancestral HMA (ancHMA) increase binding to AVR-PikD.
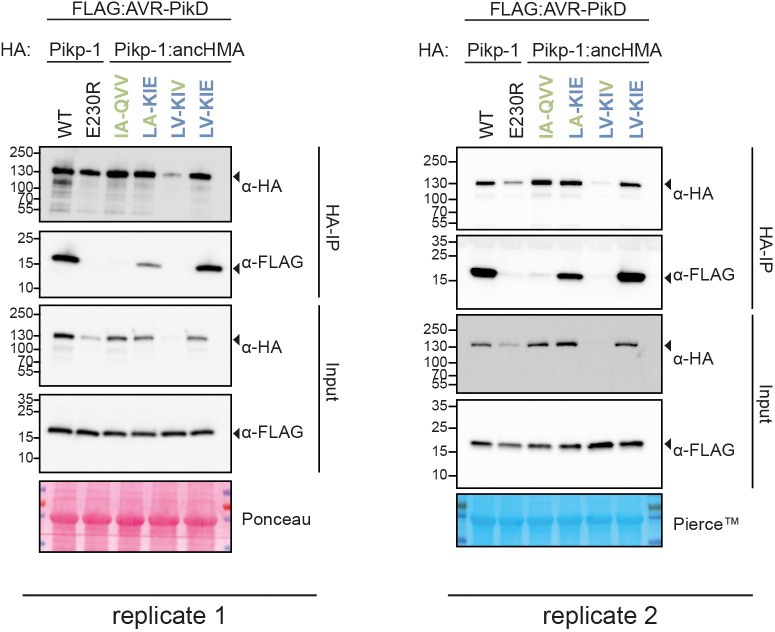
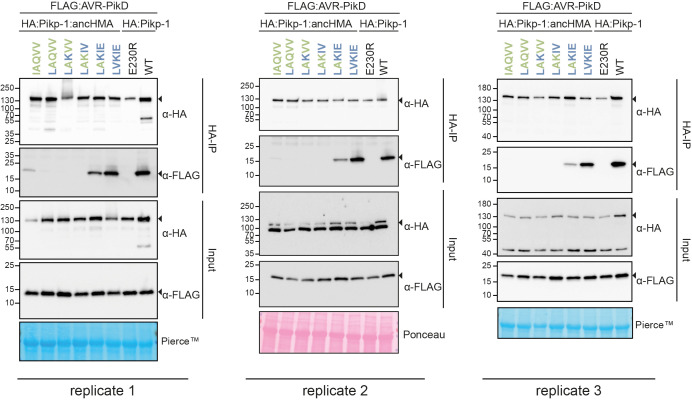
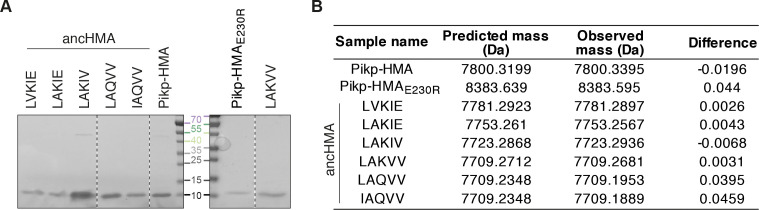
(A) Schematic representation of a neighbour joining (NJ) phylogenetic tree of the heavy metal-associated (HMA) domain from Oryza spp. (shown in Figure 3—figure supplement 2). The scale bar indicates the evolutionary distance based on the number of base substitutions per site. Historical mutations in the IAQVV/LVKIE region acquired over the course of Pikp-HMA evolution are shown next to the appropriate nodes. The mutations are colour-coded to match the ancestral (green) and present-day (blue) states. (B) Co-immunoprecipitation (Co-IP) experiment illustrating in planta association of AVR-PikD (N-terminally tagged with FLAG) with Pikp-1 and Pikp-1:ancHMA (N-terminally tagged with HA), labelled above. Wild-type (WT) HA:Pikp-1 and HA:Pikp-1E230R proteins were used as a positive and negative control, respectively. Immunoprecipitates (HA-IP) obtained with anti-HA probe and total protein extracts (input) were immunoblotted with appropriate antibodies (listed on the right). Loading control, featuring Rubisco, was performed using Pierce staining. The arrowheads indicate expected band sizes. Three independent replicates of this experiment are shown in Figure 5—figure supplement 2. (C) Plot illustrating calculated percentage of the theoretical maximum response (%Rmax) values for interaction of HMA analytes, labelled below, with AVR-PikD ligand (featuring C-terminal HIS tag) determined using surface plasmon resonance. %Rmax was normalised for the amount of ligand immobilised on the NTA-sensor chip. The chart summarises the results obtained for HMA analytes at 400 nM concentration from three independent experiments with two internal repeats. Three different concentrations of the analytes (400 nM, 200 nM, 50 nM) were tested; results for the 200 nM and 50 nM concentrations are shown in Figure 5—figure supplement 4. Average Δ%Rmax (•) values represent absolute differences between values for ‘binding’ and ‘dissociation’, calculated from the average values for each sample, and serve as an off-rate approximate. Statistical differences among the samples were analysed with Tukey’s honest significant difference (HSD) test (p<0.01); p-values for all pairwise comparisons are presented in Supplementary file 1I.