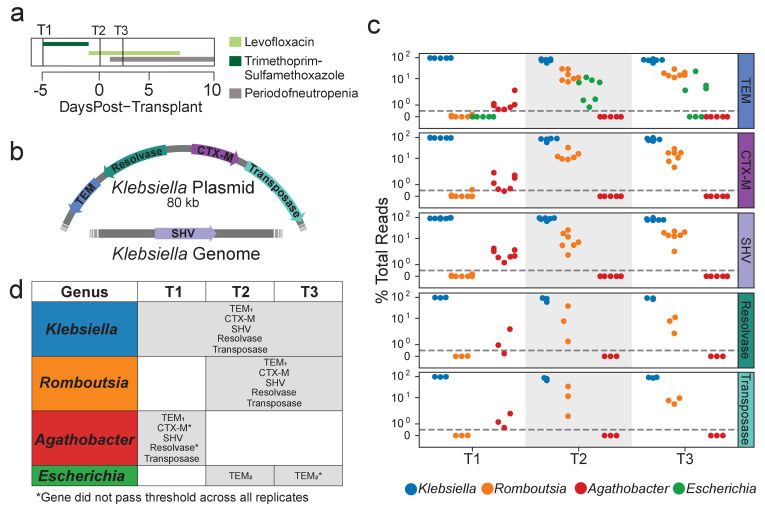
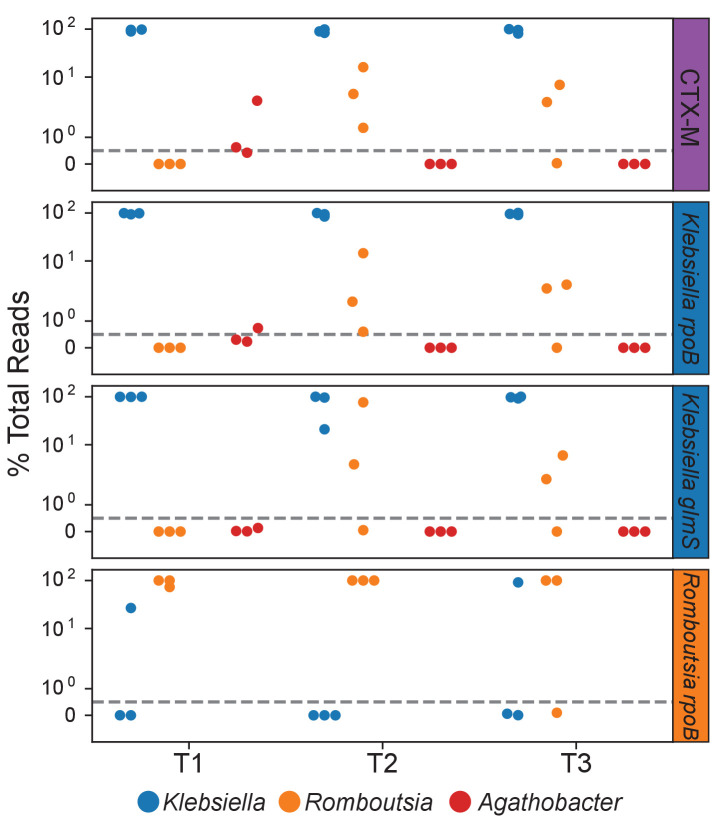
Figure 2. Extended spectrum beta-lactamase genes are associated with both pathogenic and commensal species.
(a) Summary of treatment and sample time points for patient B335. (b) Depiction of an 80 kb plasmid carried by K. pneumoniae harboring the blaCTX-M, blaTEM, Tn3 transposase and resolvase genes. The blaSHV gene is presumed to be carried within the K. pneumoniae genome. Placement of these genes was inferred from metagenomic assemblies of patient B335’s gut microbiome sample. (c) OIL-PCR results for each of the genes depicted in (a) patient B335 at three time points. For all gene-taxa associations, the percent of total OIL-PCR reads for that gene-time point is plotted. All species passing our detection threshold of 0.5% (dotted line) at any of the three time points is included in this plot. (d) A table summarizing the results in (b). All gene-taxa associations for each time point passing our detection thresholds are listed. Two SNP variants of TEM were detected and denoted with subscript numbering. Gene-taxa associations which did not consistently pass our detection threshold across all technical replicates are noted (*).