Fig. 2.
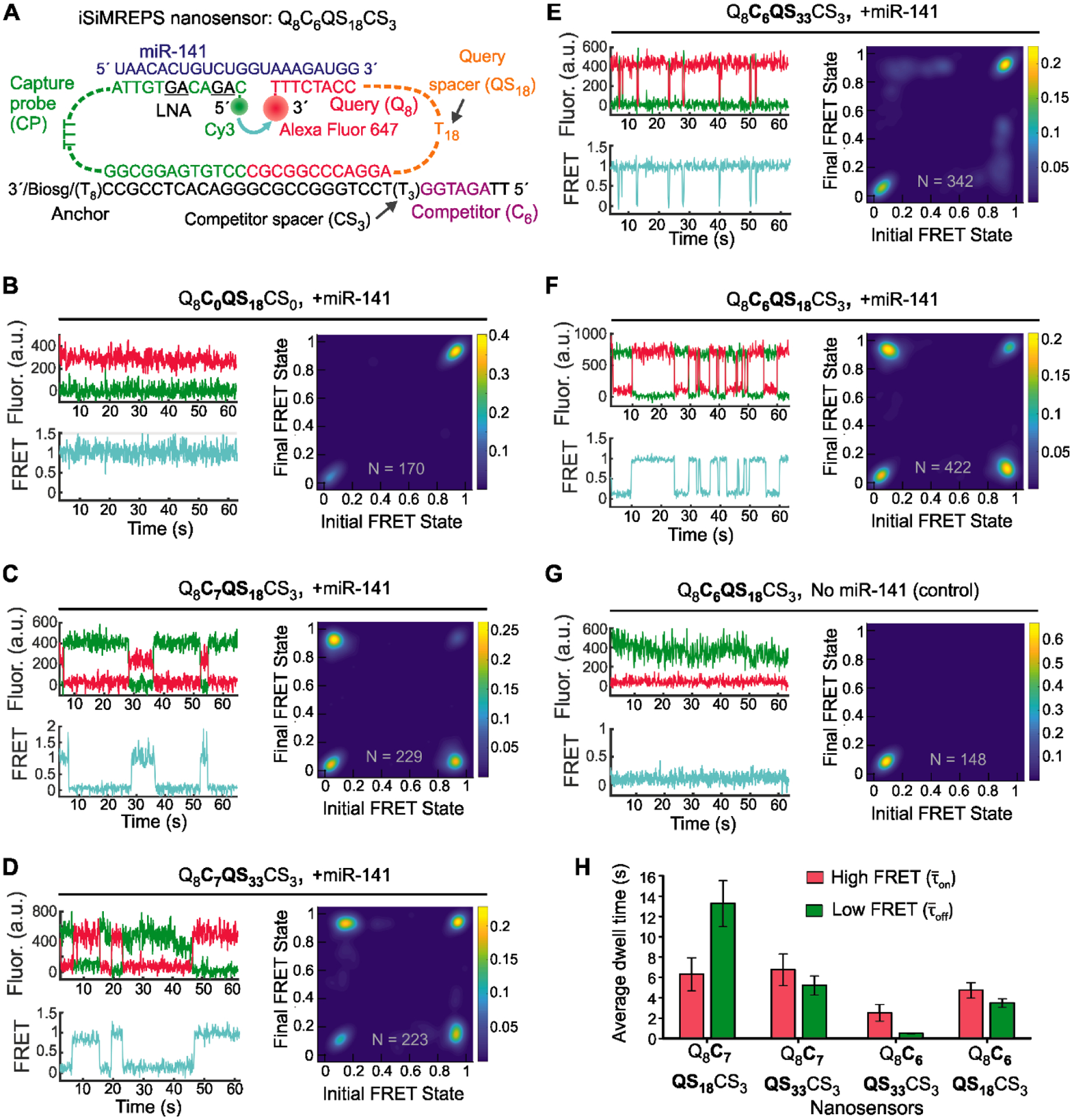
Design and optimization of iSiMREPS for detection of a miRNA. (A) Design of the optimized Q8C6QS18CS3 smFRET-based iSiMREPS sensor for detection of miR-141. The CP stably binds with the miRNA target with the assistance of locked nucleic acid residues (black and underlined) that increase the stability of the DNA-RNA duplex. The query (8 nt) switches between being bound to the 8 nt overhang of the target or to a 6 nt competitor sequence that extends from the anchor, resulting in dynamic kinetic smFRET fingerprints. (B-G) TODP plots and representative traces for different iSiMREPS sensor designs that have fixed query (8 nt), varying competitor (6 and 7 nt), fixed competitor spacer (3 nt), and varying query spacer (3, 18 and 33 nt) lengths in the presence of miR-141, as well as control without miR-141. The smFRET dynamics of each sensor is indicated. (H) The average dwell times of the high-FRET (red) and low-FRET (green) interactions for each sensor design. All data are presented as mean ± s.d., where n = 3 populations of a split data set for each condition.
