Fig. 3.
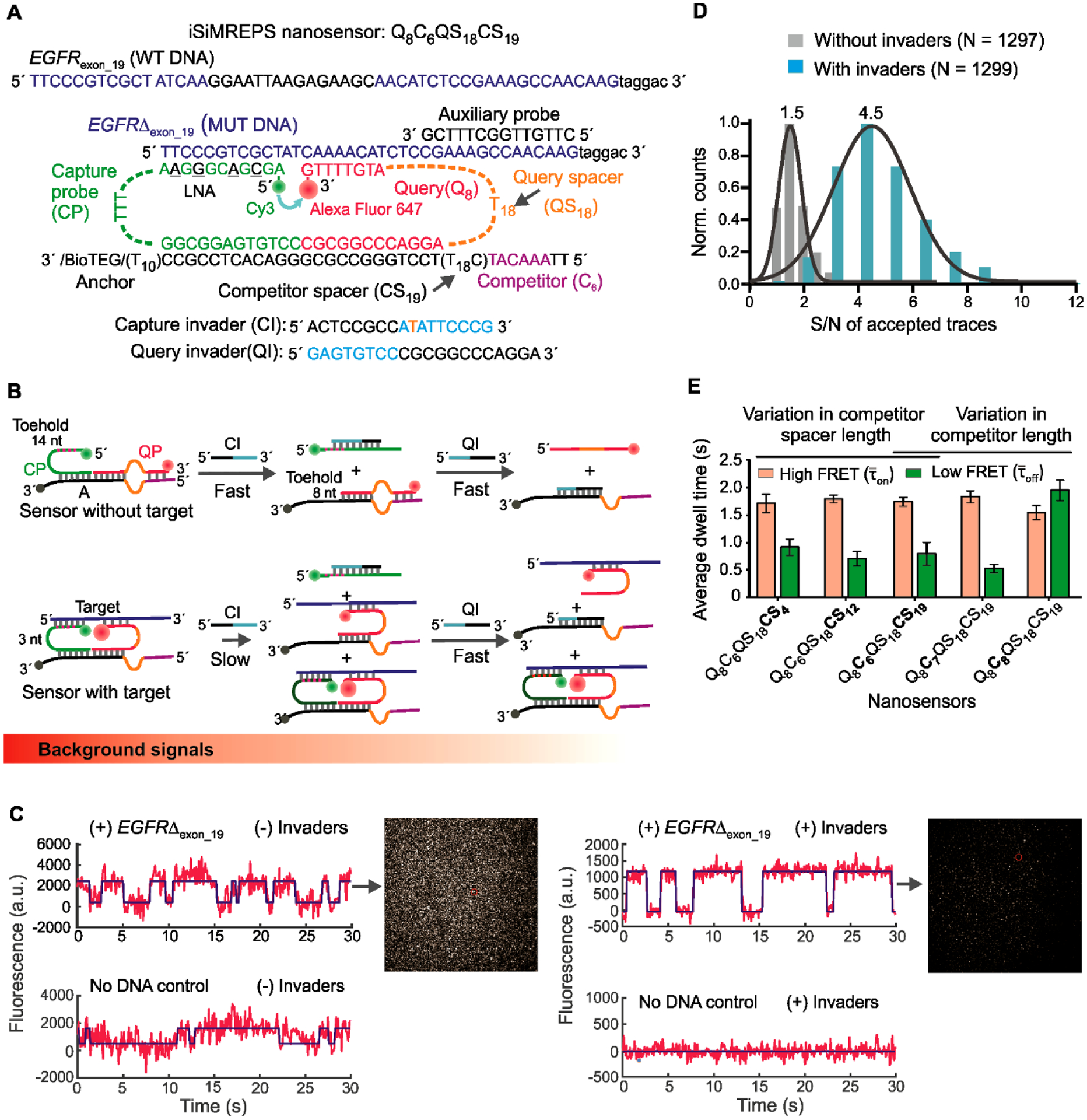
Design and optimization of iSiMREPS for detection of a ctDNA biomarker mutant DNA sequence. (A) Design of optimized smFRET-based iSiMREPS sensor for the detection of EGFR exon 19 deletion mutant DNA (EGFRΔexon_19). Two invaders, which are used to remove non-target-bound fluorescent probes from the surface, are also shown. (B) Schematic depiction of the removal of non-target-bound fluorescent probes (top) using CI and QI, and the much slower side reaction that removes target-bound probes (bottom). Each non-target-bound sensor has an exposed 9 nt toehold on the CP that binds with CI (cyan) and initiates the toehold displacement cascade. A 3 nt toehold on the CP in target-bound sensors can also bind with CI and ultimately prevent detection of a target molecule, but this reaction occurs much more slowly due to the shorter toehold. (C) Comparison of single-molecule FRET traces of iSiMREPS sensor in the presence (top) or absence (bottom) of the target sequence containing the EGFRΔexon_19. Background signals are significantly reduced with the application of invaders (right panel) compared to samples imaged without invader treatment (left panel). (D) Comparison of signal-to-noise (S/N) ratio with (cyan) and without (grey) invaders. (E) The average dwell times spent in the high-FRET (light red) and low-FRET (green) states for different iSiMREPS sensors designs. All data are presented as mean ± s.d., with n = 3 independent experiments.
