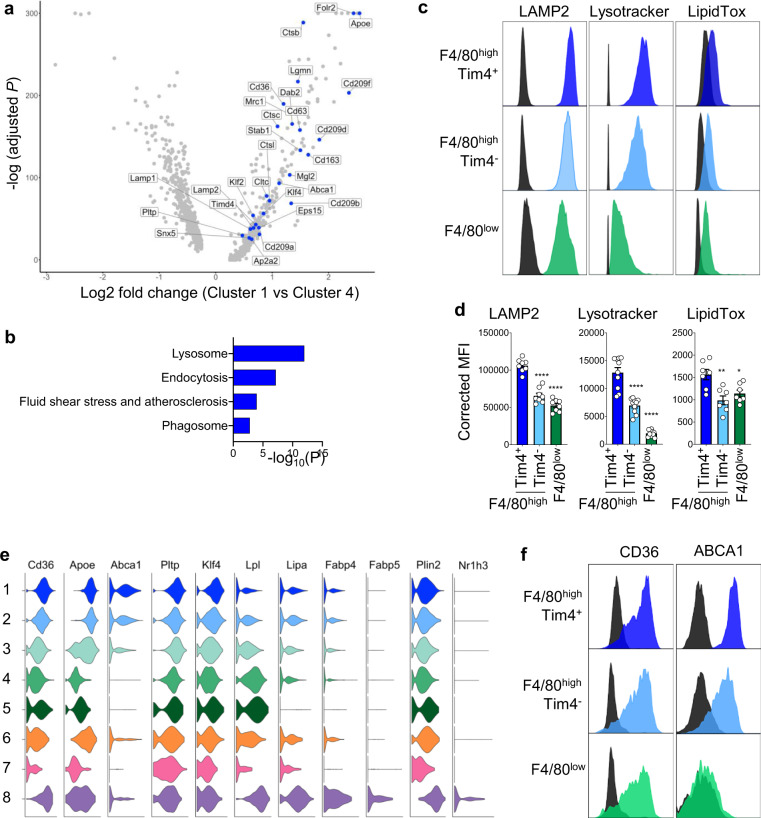
Fig. 3. Lyve1+Tim4+ ATMs have high lysosomal and lipid content.
a Volcano plot showing DEGs between Cluster 1 (Lyve1+ resident ATMs) and Cluster 4 (BM-derived ATMs). Examples of DEGs distinguishing Cluster 1 are shown in blue. b KEGG pathway analysis on DEGs distinguishing Cluster 1. c, d Flow cytometric analysis on epididymal AT showing histogram of the fluorescence intensity of LAMP2, lysotracker, and LipidTox in F4/80highTim4+, F4/80highTim4−, and F4/80low ATM populations as gated in Fig. 2a with FMO in black (c) and quantification of mean fluorescence intensity (MFI) for these staining on the indicated ATM populations (d). Data pooled from two independent experiments with n = 8 (LAMP2), n = 10 (lysotracker), or n = 7 (LipidTox) mice per group. Error bars show SEM. ANOVA with Sidak’s multiple comparisons test were applied after assessing normality using D’Agostino and Pearson Normality test. Significant differences are indicated by *P < 0.05, **P < 0.01, ****P < 0.0001. e Violin plots by cluster of the expression of genes involved in lipid metabolism. f Flow cytometric analysis showing histogram of the fluorescence intensity of CD36 and ABCA1 in ATM populations as defined in c. Data representative of n = 7 mice per group in two independent experiments.