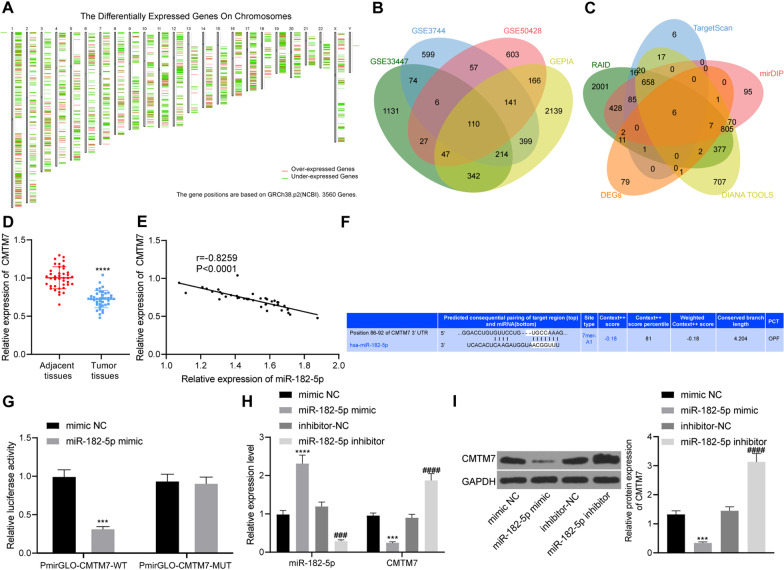
Fig. 5.
CMTM7 is a target of miR-182-5p. A A heat map of differential gene expression in breast cancer samples from the TCGA database by GEPIA (from left to right, chromosomes 1–22 and sex chromosomes are present in sequence); B Venn diagram of differential genes screened from datasets GSE33447, GSE3744 and GSE50428 in GEO database and GEPIA; C Venn diagram of miR-182-5p downstream genes and significantly differential genes predicted by RAID, TargetScan, mirDIP and DIANA TOOLS databases; D CMTM7 expression in cancer tissues and adjacent normal tissues of 40 breast cancer patients detected by RT-qPCR, he statistical power was 1; E Pearson correlation analysis of the expression of miR-182-5p and CMTM7 in breast cancer tissues, the statistical power was 0.999; F Binding site between miR-182-5p and CMTM7 predicted by the TargetScan website; G Binding of miR-182-5p to CMTM7 confirmed by dual luciferase experiment, the statistical power was 1; H Expression of miR-182-5p and CMTM7 of HUVECs detected by RT-qPCR, the statistical power was 1; I Expression of CMTM7 protein of HUVECs detected by Western blot analysis, the statistical power was 1. ***p < 0.001, ****p < 0.0001 compared with adjacent normal tissues, or HUVECs treated with mimic NC. ###p < 0.001, ####p < 0.0001 compared with HUVECs treated with inhibitor NC. The experiment was conducted three times independently