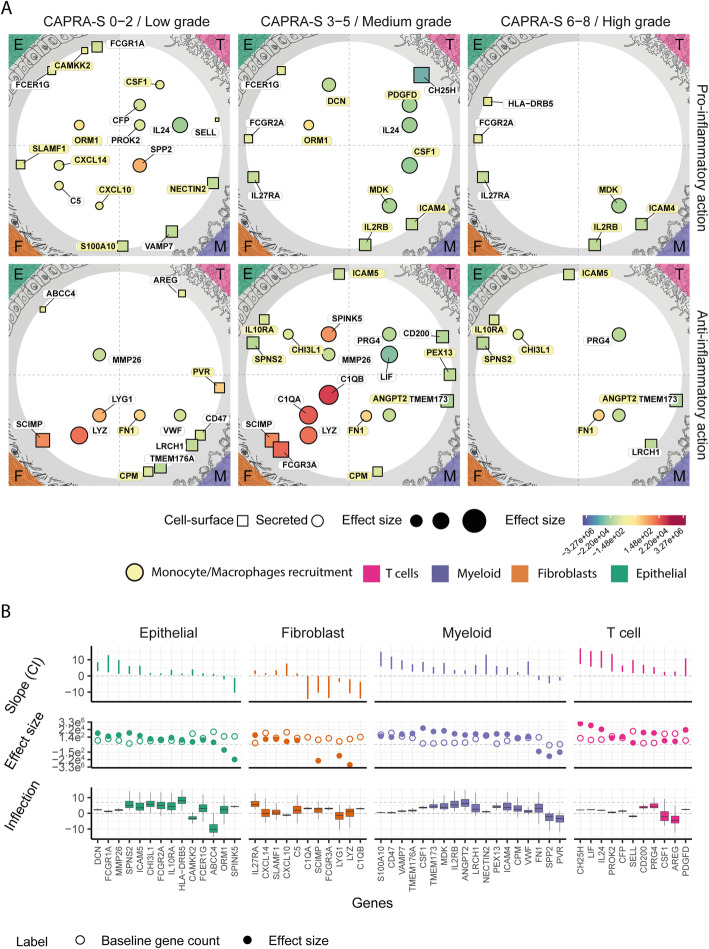
Fig. 3.
Multi cell-type immune-modulation changes with risk progression and is mainly targeted at monocyte-derived cells. The landscape of the immune-modulation related genes encoding cellular interface-proteins (i.e. cell-surface or secreted) inferred to be differentially transcribed across CAPRA-S risk scores, grouped by cell type. A Map of the secretory (represented as circles) and cell-surface (represented as squares) protein-coding genes that are differentially transcribed across the four cell types. The data point size is proportional to the baseline transcript abundance. The colour coding represents the effect size. Genes with a similar inflection point (i.e. at what stage of the disease a transcriptional change happens) are clustered vertically (CAPRA-S risk score < =2, > 2 and < =5 and > 5). Genes are split horizontally according to their pro- or anti-inflammatory role. Genes encoding for proteins that target monocyte-derived cells are highlighted in yellow. B Statistics of the differentially transcribed genes displayed in panel (A). Top: credible interval of the association between transcript abundance and CAPRA-S risk score. Middle: inferred effect size (full dots) and baseline transcription (empty dots). Bottom: credible interval of the CAPRA-S value for the transcriptomic change (i.e. inflection point; e.g., the gene HLA − DRB5 is upregulated in late stages of the disease)