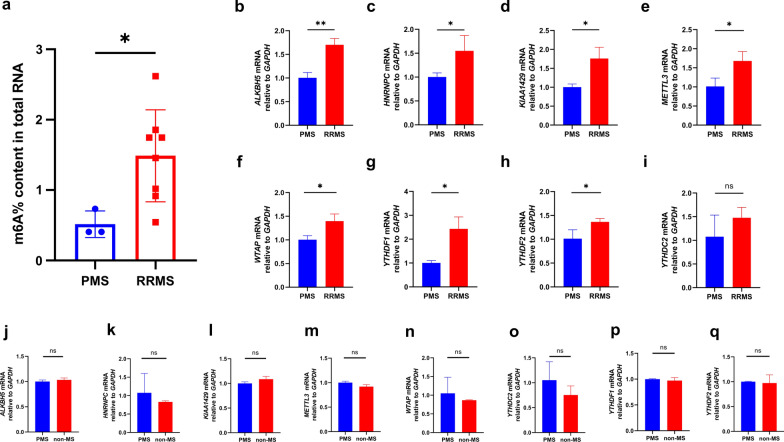
Fig. 7.
The total m6A level and qRT-PCR validation of the m6A-related feature genes in patients with MS. a The total m6A RNA methylation context of total RNA between PMS patients and RRMS patients (PMS vs. RRMS = 0.515 ± 0.154% vs. 1.488 ± 0.611%, p = 0.036). b–i The gene expression of the feature genes ALKBH5, HNRNPC, KIAA1429, METTL3, WTAP, YTHDF1, YTHDF2, and YTHDC2 between PMS patients and RRMS patients (ALKBH5: PMS vs. RRMS = 1.004 ± 0.091 vs. 1.702 ± 0.110, p = 0.002; HNRNPC: PMS vs. RRMS = 1.002 ± 0.070 vs. 1.549 ± 0.262, p = 0.046; KIAA1429: PMS vs. RRMS = 1.003 ± 0.068 vs. 1.760 ± 0.242, p = 0.013; METTL3: PMS vs. RRMS = 1.015 ± 0.178 vs. 1.680 ± 0.202, p = 0.025; WTAP: PMS vs. RRMS = 1.002 ± 0.072 vs. 1.399 ± 0.123, p = 0.017; YTHDF1: PMS vs. RRMS = 1.003 ± 0.083 vs. 2.432 ± 0.407, p = 0.008; YTHDF2: PMS vs. RRMS = 1.012 ± 0.153 vs. 1.364 ± 0.057, p = 0.038; YTHDC2: PMS vs. RRMS = 1.078 ± 0.372 vs. 1.478 ± 0.180, p = 0.243). j–q The gene expression of the feature genes ALKBH5, HNRNPC, KIAA1429, METTL3, WTAP, YTHDF1, YTHDF2, and YTHDC2 between PMS patients and non-MS patients (ALKBH5: PMS vs. non-MS = 1.000 ± 0.026 vs. 1.034 ± 0.030, p = 0.295; HNRNPC: PMS vs. non-MS = 1.075 ± 0.429 vs. 0.830 ± 0.024, p = 0.465; KIAA1429: PMS vs. non-MS = 1.000 ± 0.025 vs. 1.085 ± 0.048, p = 0.092; METTL3: PMS vs. non-MS = 1.000 ± 0.024 vs. 0.920 ± 0.034, p = 0.005; WTAP: PMS vs. non-MS = 1.052 ± 0.351 vs. 0.866 ± 0.010, p = 0.495; YTHDC2: PMS vs. non-MS = 1.051 ± 0.301 vs. 0.753 ± 0.149, p = 0.277; YTHDF1: PMS vs. non-MS = 1.000 ± 0.007 vs. 0.972 ± 0.047, p = 0.457; YTHDF2: PMS vs. non-MS = 1.000 ± 0.006 vs. 0.973 ± 0.135, p = 0.790). The significant levels were set at p < 0.05 (*), < 0.01 (**), and < 0.001 (***)