ABSTRACT
Bacteriophage serine integrases catalyze highly specific recombination reactions between defined DNA segments called att sites. These reactions are reversible depending upon the presence of a second phage-encoded directionality factor. The bipartite C-terminal DNA-binding region of integrases includes a recombinase domain (RD) connected to a zinc-binding domain (ZD), which contains a long flexible coiled-coil (CC) motif that extends away from the bound DNA. We directly show that the identities of the phage A118 integrase att sites are specified by the DNA spacing between the RD and ZD DNA recognition determinants, which in turn directs the relative trajectories of the CC motifs on each subunit of the att-bound integrase dimer. Recombination between compatible dimer-bound att sites requires minimal-length CC motifs and 14 residues surrounding the tip where the pairing of CC motifs between synapsing dimers occurs. Our alanine-scanning data suggest that molecular interactions between CC motif tips may differ in integrative (attP × attB) and excisive (attL × attR) recombination reactions. We identify mutations in 5 residues within the integrase oligomerization helix that control the remodeling of dimers into tetramers during synaptic complex formation. Whereas most of these gain-of-function mutants still require the CC motifs for synapsis, one mutant efficiently, but indiscriminately, forms synaptic complexes without the CC motifs. However, the CC motifs are still required for recombination, suggesting a function for the CC motifs after the initial assembly of the integrase synaptic tetramer.
IMPORTANCE The robust and exquisitely regulated site-specific recombination reactions promoted by serine integrases are integral to the life cycle of temperate bacteriophage and, in the case of the A118 prophage, are an important virulence factor of Listeria monocytogenes. The properties of these recombinases have led to their repurposing into tools for genetic engineering and synthetic biology. In this report, we identify determinants regulating synaptic complex formation between correct DNA sites, including the DNA architecture responsible for specifying the identity of recombination sites, features of the unique coiled-coil structure on the integrase that are required to initiate synapsis, and amino acid residues on the integrase oligomerization helix that control the remodeling of synapsing dimers into a tetramer active for DNA strand exchange.
KEYWORDS: site-specific DNA recombination, serine recombinase, synaptic complex, phage A118, Listeria monocytogenes
INTRODUCTION
Bacteriophage integrases catalyze exquisitely regulated DNA recombination reactions that are integral to the life cycles of temperate phages and can lead to symbiotic relationships with their bacterial host (1). The DNA rearrangements occur only at integrase-specific DNA sites, but these can be located on separate DNA molecules to yield intermolecular fusions or positioned on the same DNA molecule in a direct or inverted orientation to yield intramolecular deletions or inversions, respectively. Reactions are unidirectional yet are reversible under the control of an integrase-specific recombination directionality factor (RDF) (2). These properties, combined with their high reaction efficiencies both in vivo and in vitro, make these enzymes attractive for repurposing into molecular tools for genome engineering and synthetic biology (3–10). To be most useful in these applications, a comprehensive understanding of the reaction mechanism and how it is controlled is needed.
There are two major families of phage integrases, those that use a tyrosine-based and those that use a serine-based active site for DNA chemistry (11). The phage A118 integrase (Int) is a serine recombinase (SR) that is a member of the subfamily of “large” serine recombinases (LSRs) (12–14). It catalyzes the integration of the phage A118 genome at the attP locus into the attB locus located within the comK gene on the Listeria monocytogenes chromosome to generate the hybrid attL and attR sites (Fig. 1A) (15, 16). The reversal of this reaction, phage excision, requires the A118-encoded RDF Gp44 (17). Because the comK product regulates essential virulence genes during growth within macrophages, the excision of phage A118 is an important determinant of the pathogenesis of L. monocytogenes (18, 19). Other LSRs regulate terminal differentiation pathways in microorganisms, such as sporulation in various Gram-positive species or heterocyst development in cyanobacteria (20–22). Certain LSRs can mediate DNA transposition (23, 24), and some LSRs are associated with integrative and conjugative elements through which they promote the horizontal transfer of gene clusters, including antibiotic resistance determinants (25, 26). DNA elements conferring resistance to methicillin and other β-lactam antibiotics in Staphylococcus aureus are mobilized by LSRs (27, 28).
FIG 1.
The A118 integrase reactions and structural models. (A) Phage A118 integration and excision. The A118 integrase recombines the attP site on the phage genome (thick line) with the attB site in the comK gene of the Listeria chromosome (thin line) in a reaction inhibited by the phage-encoded Gp44 protein. Gp44 is required for the integrase-catalyzed excision of the prophage between the hybrid sites attL and attR to regenerate the circular extrachromosomal phage genome. (B) Schematic representation of the domains of the integrase dimer bound to the different att site sequences. The catalytic domain (cat) is connected to the recombinase domain (RD) on the opposite side of the DNA duplex through the helix E region (in brown). The zinc-containing domain (ZD), which is connected to the RD via a flexible peptide linker, can be positioned on either side of the DNA helix depending on the location of its DNA recognition sequence. DNA exchange occurs over the red GG core dinucleotide. The color-coding of the Int domains and CC motifs (red) is maintained throughout the figure. (C) Structural models of the integrase dimer bound to attP, attB, and attL. The mobile CC motifs (red) are oriented in these models as in chain C in the structure under PDB accession number 4KIS, except for the CC-paired conformation at attL. In the attL CC-paired model, the CC dimer structure, based on the structures under PDB accession numbers 5U96 for chains A and B and 4KIS for chain C, was manually positioned onto the Int-attL model. No connections were made over the unstructured peptide segments between the ZDs and the CC motifs (see also reference 31). The helix E segments are yellow in this panel to better visualize the paired CC motifs. (D) Schematic models of attB × attP and attL × attR synaptic complexes. Modeling suggests that considerable flexibility within the CC motifs is required to achieve interactions between the tips of the CC motifs of interacting integrase dimers. Gp44 (tan) binding to the base of the CC motif and ZD is proposed to be required to reorient the CC motifs in attL and attR from the paired configuration within dimers (C, bottom) to trajectories competent for interaction between dimers to achieve attL × attR synapsis (30).
Like all serine recombinases, LSRs have a relatively conserved catalytic domain followed by a long oligomerization helix (helix E). LSRs are distinguished from other SRs by having a much larger C-terminal segment consisting of two independently folded DNA-binding domains (Fig. 1B and C) (29). The recombinase domain (RD) is linked to helix E and connected to the C-terminal zinc-containing domain (ZD) by a flexible polypeptide segment. The ZD contains a long flexible coiled-coil (CC) motif that protrudes away from the DNA. The CC motif is connected to the ZD via unstructured polypeptide segments leading to its mobility. The crystal structure of an attP half-site complex with the C-terminal domain (CTD) from phage L1 Int, whose sequence is nearly identical to that of A118 Int, contains four complexes within the crystallographic asymmetric unit that each show different trajectories for their CC motifs (30). Crystal structures and biophysical properties of CC motifs in isolation have demonstrated that they interact with each other at their tips (31), and a body of data on several serine integrase systems provides evidence that this interaction controls the recombination reaction (17, 30–34).
Multiple features of LSR systems are believed to combinatorially regulate the site-specific recombination reactions (13, 14). As noted above, a distinguishing feature of this subfamily of SRs is the mobile CC motif. A118 or L1 Int mutants lacking the CC motif are completely defective in recombination in vitro, although the requirement for the CC motif is relaxed in intramolecular in vivo reactions (17, 30). The CC motif is proposed to function in the selective pairing of att sites to form the synaptic complex (SC). The relative trajectories of the CC motifs extending from att sites bound by Int dimers are proposed to be determined by the sequence architecture of the particular att site, as diagrammed in Fig. 1B and modeled in Fig. 1C (30, 35). In these models, the DNA recognition determinants for ZD binding on the 50-bp attP site are extended by a half-helical DNA turn relative to the 40-bp attB site, which will position the CC motifs on opposite sides of the two complexes. These trajectories enable the CCs on Int dimers bound to attB and attP to interact at their tips (Fig. 1D) (31). The binding of the RDF (Gp44) to the base of the CC motif and to a second region on the C-terminal domain is proposed to orient the CC helices of Int dimers bound to attL and attR appropriately for synapsis (Fig. 1D, bottom) (17, 30). Without Gp44, the CC helices from each subunit bound to attL or attR can associate, thereby potentially locking the dimers into an inactive conformation (Fig. 1C, bottom). Gp44 also inhibits attB × attP recombination, presumably by directing the CC motifs into a configuration that is incompatible with functional synapsis.
The helix E region has been shown to be critical for regulating synapsis in small SRs known as serine resolvases and invertases (36–43) and is likely to be important for LSRs. During the process of stable SC formation by small SRs, DNA-bound synapsing dimers merge into a compact tetramer that becomes competent for cleavage and exchange of DNA strands by subunit rotation (44, 45). The dimer-to-tetramer conversion is the key control step limiting recombination by small SRs (46–48). This massive conformational rearrangement involves the remodeling of the helix E regions of the original dimers into synaptic dimer and rotating dimer interfaces within a tetramer. The only structure available of a serine integrase catalytic domain is that of a partly isomerized tetramer of phage TP901 Int that is not in an active conformation for DNA exchange (49).
The final determinant controlling directionality is the orientation of the crossover dinucleotides at the center of the recombination sites (16, 32, 50–54). The att sites must be aligned correctly for the cleaved asymmetric core sequences, GG for A118, to ligate after subunit rotation and generate recombinant products. For attP × attB reactions, the crossover dinucleotide is the sole determinant of site polarity (16, 50, 51).
In the present work, we first directly test the att site architecture hypothesis postulated by Van Duyne and coworkers to specify the identities of the serine integrase att sites (30). Our analyses of reconstructed A118 att sites provide compelling evidence that the trajectories of CC motifs are indeed determined by the att site architecture and that the RDF Gp44 regulates the CC trajectories. We then define molecular aspects of the CC motifs that are important for controlling synapsis, including the importance of CC motif length and the finding that some amino acid residues near the tip may differentially participate in attB × attP versus Gp44-activated attL × attR reactions. We show that the helix E region plays an important role in regulating recombination by identifying key residues on helix E that control synaptic complex formation. One of the helix E mutants facilitating synapsis overrides the requirement for the CC motifs, resulting in promiscuous synapses. Other helix E mutants still require the CC motifs, demonstrating the functional interplay between the two regions for directional synapsis. The recombination properties of the CC-independent synapsing mutant suggest a continuing role for the CC motifs after initial synapsis.
RESULTS
Sequence architecture defining A118 att sites.
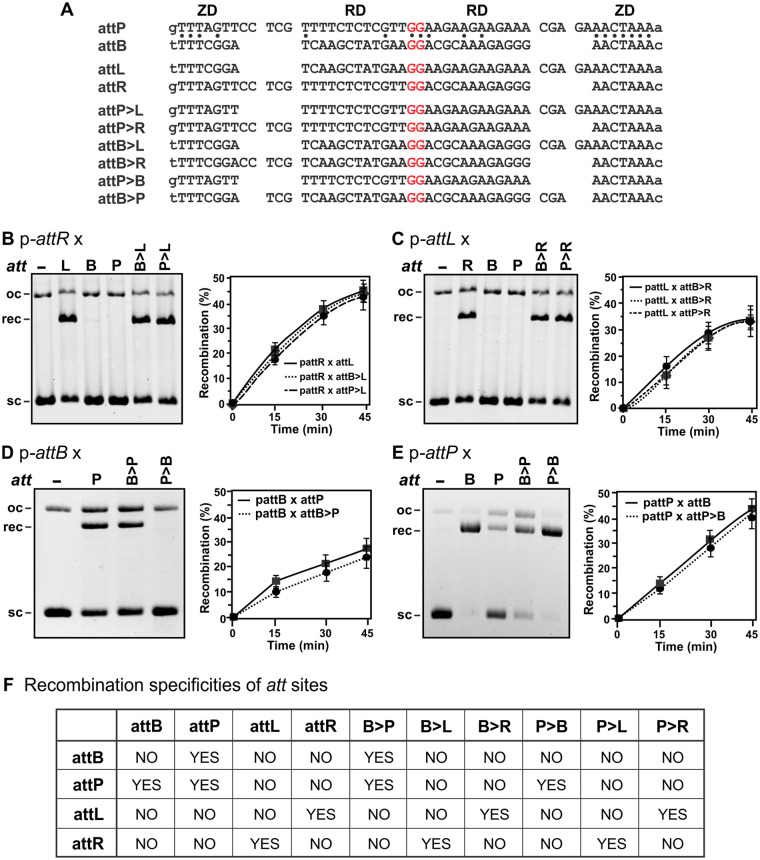
We first directly tested the model that the spacing between the DNA recognition determinants for the RD and ZD is the key feature that specifies att site identity (30). In this model, the RD and ZD recognition sequences in each attB half-site are 5 bp closer to each other than in attP, and the hybrid attL and attR sites reflect these spacing differences (Fig. 2A). We asked if the attP sequence could be converted to functional attL and attR sites by deleting 5 bp between the 7-bp segment bound by the ZD and the 26-bp region surrounding the central GG crossover dinucleotide on the left and right half-sites, respectively. The 19-bp segment to the left of the crossover GG in the reconstructed attP>L site shares only 6 bp in common with wild-type (wt) attL. Nevertheless, attP>L recombines with attR as effectively as attL in Gp44-dependent reactions (Fig. 2B). The reconstructed attP>R site, which shares only 4 of 12 bp over the RD-binding sequence to the right of the GG crossover, also efficiently recombines with attL when Gp44 is present (Fig. 2C). Likewise, the attB site was efficiently converted to functional attL or attR sites by adding the 5-bp sequence from attP between the RD core and ZD-binding sites, respectively (Fig. 2A to C). For the reconstructed attB>R site, 11 of 24 bp over the left half-site are identical to those of wild-type attR (Fig. 2A).
FIG 2.
Reconstructed att sites. (A) DNA sequences of the native attP, attB, attL, and attR sites and those reconstructed from attP and attB by changing the spacing between the ZD and RD recognition determinants. Red bases are the core nucleotides, and base identities between attP and attB when aligned as shown are marked. (B to E, left) Gels representing products of 20-min Int reactions between 100-bp DNA fragments containing wt and reconstructed att sites and supercoiled (sc) plasmids containing attR (B), attL (C), attB (D), and attP (E). oc is open-circular plasmid. Reaction mixtures for panels B and C included Gp44. The recombinant product (rec) is a linear plasmid (Fig. 1A). (F) Summary table of the recombination specificities of wt and reconstructed att sites.
In addition, attP was reconstructed into a functional attB site and attB was reconstructed into a functional attP site by the deletion or addition, respectively, of the 5-bp linker region on both half-sites (Fig. 2A). attB>P recombined with attB almost as well as wild-type attP (Fig. 2D). attB>P also exhibited significant recombination with attP, as is observed with wild-type attP × attP reactions by A118 Int (Fig. 2D). attP>B exhibited recombination with attP equally as efficient as that of the wild-type attB site (Fig. 2E). The specificities of each of the four reconstructed att sites parallel those of the native sites (Fig. 2F).
Gp44 binding changes the shape of Int-bound complexes with reconstructed att sites.
We showed in a previous report that Int+Gp44 forms electrophoretically distinct complexes with different att sites (17). For example, the binding of Gp44 to Int-attB complexes increases the mobility of the DNA complexes through polyacrylamide relative to attB complexes containing only Int, even though the Gp44-bound complexes have a higher mass. In contrast, Gp44-bound Int complexes with attL, attR, or attP migrate slower to various extents relative to their respective Int-only complexes. The differences in mobility induced by Gp44 binding most likely reflect the relative trajectories of the CC motifs on the shape of the complex as it migrates through the gel. In all cases, the binding of Gp44 to Int on reconstructed att sites results in similar migrations to the respective wild-type sites (Fig. 3). These include the attB site reconstructed from attP (attP>B) and attB sites converted into other att sites (attB>L and attB>R), which are the most diagnostic.
FIG 3.
Electrophoretic mobilities of Int and Int+Gp44 bound to reconstructed att sites. The binding of the Int dimer shifts the mobilities of the DNA fragments. Gp44 binding further retards Int complexes on attP, attL, and attR but increases the electrophoretic mobility on attB complexes, even though the mass is increased by 17 kDa. The differences in migrations are believed to reflect the architectures of the different complexes, specifically the trajectories of the CC motifs.
The recombination activities and specificities of the reconstructed att sites, combined with the electrophoretic migrations of att complexes, provide strong support for ZD-RD spacing solely determining att site functional identity. The underlying mechanism is likely to be the att-site-specific trajectories of the CC motif emanating from the ZD on the DNA complex, which is altered upon Gp44 binding.
Features of the CC motif important for recombination. (i) The length of the CC motif is important for recombination.
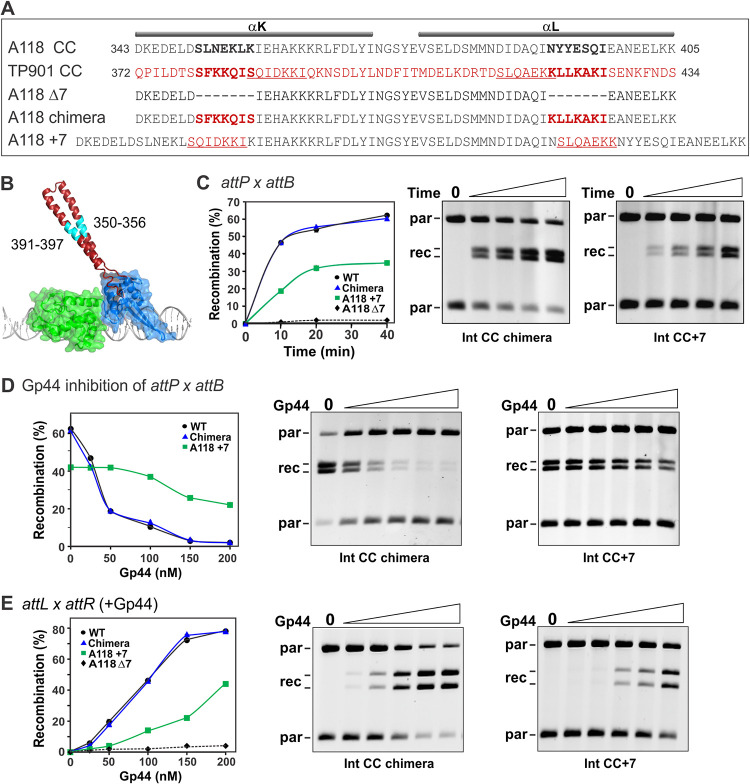
Residues within the tips of the CC motifs have been shown to be essential for recombination in vitro and are proposed to function in att site synapsis (17, 30–34). A minimum length of the CC arms would be expected to be essential in order for the tips to associate. To evaluate whether the length of the CC motif is important, an Int mutant was constructed that removed two α-helical turns from the centers of each of the helices within the CC motif (helix K residues 350 to 356 and helix L residues 391 to 397) (Fig. 4A and B). This mutant, A118 IntccΔ7, did not support attP × attB or attL × attR (+Gp44) recombination in vitro (Fig. 4C and E). We replaced the missing residues within the A118 IntccΔ7 mutant with residues from the analogous segments of the TP901 serine integrase to generate A118 IntCC-chimera. Only 1 of the 7 substituted residues on each CC helix of this mutant is identical to the native A118 sequence (Fig. 4A). Nevertheless, A118 IntCC-chimera catalyzed attP × attB recombination as well as wild-type Int (Int-wt) (Fig. 4C). Moreover, attP × attB recombination by IntCC-chimera was inhibited by Gp44, in a manner similar to that of Int-wt (Fig. 4D), and Gp44-dependent attL × attR recombination by IntCC-chimera was as efficient as that of Int-wt (Fig. 4E).
FIG 4.
The length of the CC motif is important for activity. (A) Alignment of the amino acid residue sequences of the CC segments from the A118 and TP901 integrases. Below are the sequences of the CC length mutants. (B) Structure of the L1 integrase RD (green) and ZD (blue) rendered as a transparent surface. The CC motif (PDB accession number 4KIS for chain C with the tip appended from PDB accession number 5U96 for chain A) is highlighted in red; cyan denotes the segment deleted in A118 CCΔ7 or replaced with TP901 residues (A118 chimera). (C) attP × attB deletion reactions by the wt and CC mutant integrases. On the left are plots of representative reaction time courses, and on the right are gel images of reactions by the Int CC chimera and Int CC+7 mutants. The DNA was digested with NdeI, which cleaves the parental (par) plasmid twice and linearizes the product circles, prior to agarose gel electrophoresis (see Fig. S4 in the supplemental material). (D) Gp44 inhibition of attP × attB deletion reactions by wt and mutant integrases. Reactions were performed as described above for panel C for 20 min in the presence of 0, 25, 50, 100, 150, and 200 nM Gp44, and example data are shown. (E) Gp44-activated attL × attR deletion reactions by the wt and CC mutant integrases. Reactions were performed as described above for panel D.
We also constructed an A118 Int mutant in which the CC motifs are extended by two helical turns by adding 7 residues from TP901 Int to helices K and L (Fig. 4A). The resulting mutant (A118 Intcc+7) was partially active in catalyzing both attP × attB and attL × attR (+Gp44) recombination (Fig. 4C and E). The less efficient attP × attB recombination by Int containing the longer CC motifs could be influenced, in part, by enabling inhibitory intramolecular interactions when the Int dimer is bound at attB. Interestingly, attP × attB recombination by A118 Intcc+7 was only slightly inhibited by Gp44 (Fig. 4D), even though Gp44 binds normally to the mutant protein (not shown).
Taken together, these results demonstrate that a minimal length of the CC motif is critical for function, which is consistent with a role of the motif in the initiation or stabilization of synapsis. Int proteins containing CC motifs that are extended by 2 additional helical turns remain partially active, consistent with a flexible coiled-coil structure. This flexibility may enable tip interactions by the CC+7 motifs during attP × attB reactions even in the presence of Gp44, which for wild-type-length CC motifs is believed to preclude productive interactions. These results also demonstrate that the identities of the 7 residues within the center of the CC helices are not important for recombination or Gp44 activity, implying that their role is solely to extend the length of the CC. As expected, an Int mutant deleted for only one side of the CC (helix K residues 350 to 356) is completely defective for recombination in vitro (not shown).
(ii) Residues within the CC tip region important for recombination.
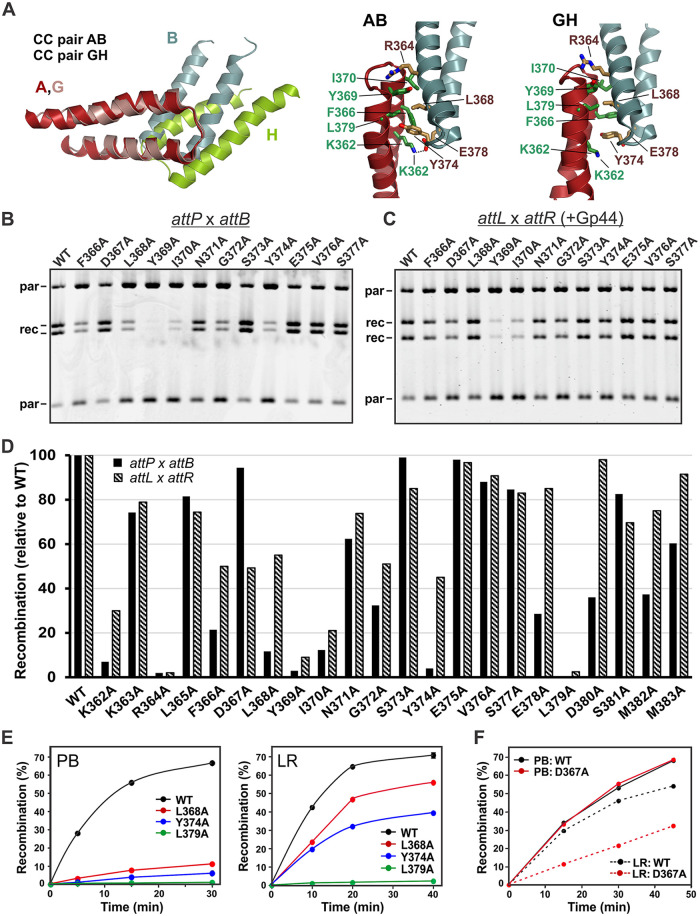
The 10 pairs of CC dimers (6 unique CC pairs) represented in the 3 crystal structures described previously by Gupta et al. (31) broadly separate into two structurally distinct classes (Fig. 5A; see also Fig. S1 in the supplemental material). When aligned over one CC unit, the partner units in the two classes exhibit about a 30° rotational difference. Moreover, there are considerable differences in the positions and interactions of side chains within the CC interfaces, even within a class. We individually replaced each of the residues over the interface region between Lys362 and Met383 with alanine to evaluate which residues are important for promoting Int recombination and to ask whether there may be differences with respect to how the CC units interact to promote attP × attB (PB) versus attL × attR (LR) recombination. Each alanine-substituted Int protein was purified and quantitatively assayed in vitro for intramolecular PB and Gp44-activated LR recombination. Representative data are shown in Fig. 5B and C, and recombination efficiencies normalized to those of wild-type Int are plotted in Fig. 5D. Our data complement and extend the mutant data of Gupta et al. (31), which evaluated PB recombination by a subset of alanine substitutions primarily in vivo.
FIG 5.
Alanine-scanning mutagenesis of residues over the tip of the CC motif. (A) Structure of CC pairs over their tip regions from the structure under PDB accession number 5U96. On the left, the AB and GH pairs were aligned over chain A (red) and chain G (pink) to highlight the rotational differences between interacting chain B (silver) and chain H (green), respectively. On the right are the AB and GH CC pairs with selected interacting side chains shown to illustrate differences in the interfaces. The ion pair between Lys362 and Glu378 is denoted on CC pair AB, but it is not present in CC pair GH. See Fig. S1 in the supplemental material for a rotated (end-on) view of the interface highlighting differences. (B) Representative intramolecular attP × attB deletion reactions by a set of CC alanine mutants. (C) Representative intramolecular Gp44-activated attL × attR deletion reactions by the same set of CC alanine mutants. Reactions in panels B and C were done for 20 min. (D) Bar graph depicting the relative reaction efficiencies for attP × attB and attL × attR reactions by each mutant. Percent recombination values from 2 to 4 deletion reactions (20 min) were averaged and scaled relative to Int-wt. (E) Representative time courses of intermolecular fusion reactions by Int-wt and selected mutants showing very low recombination rates for both attP × attB (left) and attL × attR (right) (Int-L379A) or poor recombination for attP × attB in comparison to attL × attR (Int-L368A and -Y374A). (F) Representative time courses of attP × attB and attL × attR intermolecular reactions by Int-wt and Int-D367A, which is more defective for attL × attR reactions.
The mutant data implicate 14 residues, which when replaced with alanine exhibit >2-fold decreases in PB and/or LR recombination. The positions of these residues add experimental support for the front-to-back arrangement of interacting surfaces between the CC motifs, as observed in the X-ray structures (31). Some of the residues that exhibit particularly strong mutant phenotypes may also contribute to helix-helix interactions (e.g., Tyr369 and Leu379) and to the conformation of the loop connecting the helices (e.g., Gly372) within an individual CC unit. The Arg364 side chain seems to be particularly important as the alanine substitution nearly abolishes both PB and LR recombination. Arg364 caps one end of the CC pair interfaces by engaging in multiple contacts with the critical Tyr369 plus other residues near the top of the partner CC, but the molecular details of the interactions in the two structural classes vary considerably (Fig. 5A and Fig. S1).
Significantly, a number of CC mutants exhibited differences with respect to PB versus LR recombination, with the PB reaction typically being compromised to a greater extent (Fig. 5D). Among these are K362A, F366A, L368A, Y374A, E378A, and D380A; Fig. 5E gives example time courses for intermolecular recombination reactions by L368A and Y374A, showing very low rates for PB recombination but intermediate rates for LR recombination relative to Int-wt. Some of these side chains adopt very different configurations in the CC pair structures. For example, the F366 ring is oriented into the center of the interface of the GH pair, being positioned between Tyr374 and Leu368 and abutting Leu379, whereas F366 is not contacting any of these residues in the partner CC in the AB pair (Fig. 5A and Fig. S1). In contrast, the side chains of Lys362 and Glu378 are closely bonded in CC pair AB, but the two side chains are far apart in CC pair GH (Fig. 5A). PB recombination is substantially lower than LR recombination for both K362A and Glu378A. On the other hand, D367A exhibits nearly wt PB recombination rates, but initial LR rates are 3-fold lower (Fig. 5F). The Asp367 side chain is positioned to hydrogen bond with Tyr369 in CC pair GH but not in the AB pair. The differences in the activities of these mutants, together with differences in the molecular interactions, suggest that the CC interfaces responsible for promoting PB and LR recombination may not be identical.
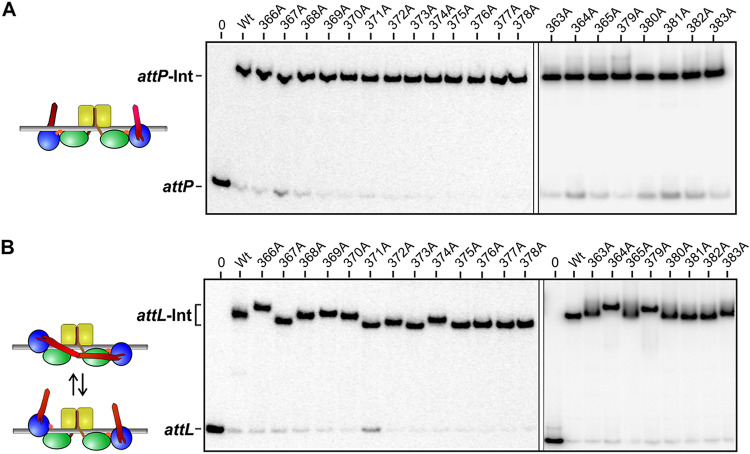
The shape of attL-Int dimer complexes is altered by CC tip mutants.
As described above and by Mandali et al. (17), the electrophoretic migrations of Int-att complexes reflect the shape of the specific att site complex as manifested by the trajectories of the CC motifs. In Int-attP complexes, the CC motifs are not predicted to interact with each other, and thus, mutational changes within the CCs are not expected to have an effect on the migrations of the complexes. As expected, the migrations of attP complexes containing Int dimers with the different alanine substitutions are unaffected (Fig. 6A). The CC motifs of Int dimer complexes bound to attL and attR are postulated to be associated with each other in an inhibitory conformation, thereby forming a more compact complex structure (30). We find that a subset of CC mutant Int dimers bound to attL migrate distinctly slower than those formed by Int-wt (Fig. 6B and Fig. S2A). Most prominent among the members of this subset are R364A, F366A, L368A, Y369A, I370A, Y374, and L379A, which correspond to those exhibiting the largest recombination defects. The slower migrations by these mutants imply a less compact structure whereby the CC motifs are less frequently associated with each other. Where tested, similar electrophoretic migration patterns were observed for complexes on attR (not shown), as expected, due to the structural similarity with attL complexes. The binding of the RDF Gp44 is proposed to disrupt the intramolecular CC contacts within Int dimers bound to attL or attR; therefore, the migration differences by Int-CC mutant complexes with attL are predicted to be abrogated when Gp44 is cobound. For all mutants tested, including those showing altered migrations without Gp44, Int-attL complexes formed in the presence of Gp44 migrated indistinguishably from Gp44-bound Int-wt complexes (Fig. S3).
FIG 6.
Electrophoretic migrations of CC alanine mutants bound at attP and attL. Int CC mutants were bound to 100-bp DNA fragments containing attP (A) or attL (B) and subjected to native PAGE (37.5:1 polyacrylamide-bisacrylamide) (Fig. S2 in the supplemental material shows examples at a 59:1 acrylamide ratio). Slow migrations by CC mutants relative to Int-wt observed on attL correlate with defective recombination and imply a less compact structure that is consistent with the CC motifs in a more open configuration. The binding of Gp44 to mutant Int-attL complexes abrogates the mobility differences (Fig. S3).
In summary, the gel migration data provide physical evidence supporting the conclusion that the CC tip mutations directly affect CC-CC interactions and that CC motifs from the subunits of dimers bound to attL (and attR) are associating with each other without Gp44 but not in the presence of Gp44. McEwan et al. reported evidence for CC interactions between subunits of ϕC31 Int dimers bound at attB (34), but gel migration experiments provide little support for CC interactions within A118 Int dimers bound at attB (Fig. S2B). The CC motif region of ϕC31 Int is predicted to be longer than that in A118 (14), which may explain this difference.
Gp44-independent attL × attR recombination by Int-CC tip mutants.
CC tip mutants that disrupt inhibitory intramolecular dimer interactions at attL and attR but retain sufficient affinities to promote synapsis potentially could enable some Gp44-independent LR recombination. We therefore tested each of the alanine substitutions for their ability to support Gp44-independent LR recombination in vitro, where product formation by Int-wt is completely dependent upon the RDF. Detectable LR recombinant products were observed only for F366A (Fig. S4A), but the Gp44-independent LR reaction was very poor. Other amino acid substitutions at residue 366 were evaluated for Gp44-independent LR recombination. F366K exhibited a similarly low efficiency as F366A, and F366L gave a trace of products; F366M/Y/W gave no detectable activity (Fig. S4B). We conclude that efficient CC tip interactions between synapsing dimers are of dominant importance for LR recombination and that destabilization of intramolecular CC interactions within dimers bound to attL and attR is insufficient for LR recombination in vitro by A118 Int.
Gupta et al. reported that several CC tip mutants of L1 Int gave weak RDF-independent LR recombination in vivo, with F366A being the strongest (31). Rowley et al. isolated the mutation Y475H within the CC-pairing region of ϕC31 Int in a genetic screen for mutants able to support weak RDF-independent LR recombination in vivo; this mutant was not active for LR and exhibited low PB recombination in vitro (32). Tyr475 in ϕC31 Int is predicted to correspond to Tyr369 in A118/L1 Int where an alanine substitution severely reduces PB and LR recombination (31; this study). The ϕC31 Int mutant E449K exhibits robust RDF-independent LR recombination in vivo and in vitro. This substitution is located at the base of the CC motif and is believed to affect the trajectory of the CC motif emanating from the ZD (32, 55).
Role of helix E residues in controlling synapsis.
A set of residues in the helix E regions and catalytic domains of small SRs have been shown to control the remodeling of dimers into synaptic tetramers, which is the key step regulating SC formation by these enzymes (46–48). We wondered if similar principles operate for the LSRs that uniquely contain CC motifs and, if so, how these distinct structural units cooperate to regulate the recombination reaction.
Wild-type A118 Int forms few, if any, SCs that are detectable by gel electrophoresis under a variety of conditions (e.g., see Fig. 7B). These include reaction mixtures containing different divalent cations, EDTA, or alcohols like glycerol or ethylene glycol; the use of catalytically defective mutants; or DNA modifications (nicks or phosphorothioates) around the DNA exchange site, which have been found to stabilize or irreversibly trap SCs in various SRs (50, 56, 57). There have been extensive studies on gain-of-function mutants of small SRs, which have often been identified through genetic screens. Many of these mutants are hyperactive for recombination and both alleviate the requirement for required cofactors and accumulate SCs, which are not detectable in the respective wild-type reactions. Using the wealth of data from the small SR systems, we introduced specific amino acid substitutions into the A118 Int catalytic and helix E regions, and the resulting mutant proteins were purified and screened for SC formation by gel electrophoresis. Of the 17 positions where one or more substitutions were evaluated, changes at 5 residues (Phe113, Met117, Gly119, Leu121, and Leu127), which were all within the helix E region (Fig. 7A and H), were found to accumulate substantial amounts of SCs. As shown in Fig. 7A, these positions are associated with hyperactivating mutations in small SRs, even though the identities of the residues vary considerably. The properties of these mutants are described below, and the structural bases for their gain-of-function activities are considered in Discussion.
FIG 7.
Helix E residues controlling synaptic complex formation. (A) Sequence of the N-terminal segment of the E helices of SRs over the region in contact with their catalytic domain or partner helix E. Blue-shaded residues are the positions of gain-of-function mutations in A118 integrase (this work), Hin DNA invertase (38, 39, 43), Tn3 or γδ resolvase (37, 41), Sin resolvase (42), and ϕC31 Int (61) that enhance SC formation. Three LSR sequences are also given with gray shading, indicating identities with the A118 integrase: a putative conjugative transposon recombinase from Clostridium difficile used for structural modeling (PDB accession number 3G13) (see panel H), the TP901 integrase, and the more distantly related ϕC31 integrase. (B) SC formation by A118 mutants. Reactions with the different A118 proteins employed 100-bp 32P-labeled attB and excess 60-bp attP fragments. Proteins in the right panel have their CC motifs deleted. (C) Migration of SCs formed by Int-M117I and -L127V with 100-bp 32P-labeled attB and excess 60- or 200-bp attP fragments. (D) Specificity of att site synapsis by A118 mutants. Example gels are given for Int-M117I and -L121I reactions with 100-bp 32P-labeled attB and excess 60-bp attB, attP, attL, and attR fragments in the absence and presence of Gp44. Note that Gp44 binding (*) to the Int dimer on attB shifts the complex to a faster-migrating species. A complete set of specificity gels is provided in Fig. S5 in the supplemental material. (E) Summary of att site synapsis specificities by the A118 integrase mutants. +f indicates a higher-mobility complex that is missing the attP DNA. (−) indicates not detected but that a higher-mobility SC could be obscured by the Gp44-bound dimeric complex (but see Fig. S5B). (F) SEC of Int-M117 without and with incubation with attP DNA. The trace of the DNA only (60-bp attP) and peaks of MW standards (catalase, 232 kDa; aldolase, 158 kDa; BSA, 67 kDa) are shown. AU, arbitrary units. (G) Recombination frequencies of wt and mutant A118 integrases with or without their CC motifs. Results are the averages and standard deviations from 20-min attB × attP deletion reactions relative to the wt (0.556 ± 0.033 deletions/substrate molecule). (H) Structural model of the A118 integrase dimer over the catalytic domain and helix E regions highlighting the locations of the SC-forming mutations in helix E. The catalytic domain of one subunit is rendered as a surface with its helix E as a dark green ribbon and the native side chains of the mutant residues in red (Cα red sphere for Gly119). The model is based on the structure under PDB accession number 3G13 (see Materials and Methods).
SCs were identified from reactions in which mutant Int proteins were incubated with 32P-labeled attB plus 4- to 5-fold-larger amounts of unlabeled attP fragments (Fig. 7B). Labeled complexes migrating slower than the dimer-attB complex were observed for A118 Int mutants F113Y, M117I, G119Y, L121I, and L127V. The formation of these complexes was enhanced by high att DNA concentrations (≥25 nM) and 10 to 30% ethylene glycol in the reaction buffer, and the SCs were stabilized during electrophoresis by low temperature and glycerol in the polyacrylamide gel. The presence of two DNA fragments in the radiolabeled SC was confirmed for Int-M117I, whereby complexes formed with labeled 100-bp attB fragments and unlabeled 60- or 200-bp attP fragments migrated increasingly slower, respectively, reflecting their different DNA masses (Fig. 7C). Complexes formed by L127V migrated faster than the SCs formed by the other 4 mutants (Fig. 7B), albeit small amounts of similarly migrating complexes were occasionally observed for the other mutants. The faster-migrating complexes generated by L127V did not contain a second DNA fragment, as shown in experiments employing different-length attP segments (Fig. 7C), but their formation depended upon the addition of the attP fragment to the reaction mixture. We believe that these complexes have an Int tetramer bound to a single 32P-labeled attB fragment and that the second att site DNA (unlabeled attP) was lost during the dimer-tetramer remodeling reaction forming the synaptic complex or during gel electrophoresis. An alternative explanation is that Int bound to labeled attB captured a second unliganded dimer from solution or that L127V is a tetramer in solution. These possibilities seem less likely because the formation of the high-mobility SC by L127V is dependent upon the presence of the attP fragment, and size exclusion chromatography (SEC) revealed L127V to be primarily dimeric.
Each of the helix E mutants was coupled with the CC motif deletion and assayed for SC formation (Fig. 7B, right). Most of the mutants were unable to form attB-attP SCs without their CC motifs. However, M117I-ΔCC efficiently formed SCs, and G119Y-ΔCC formed a small amount of SCs. The dependence on the CC motifs for SC formation by F113Y, L121I, and L127V and enhancement by G119Y provides direct evidence for the function of the CC motifs in initiating or stabilizing synapsis.
We asked whether the helix E mutants exhibited att site specificity for SC formation that reflected recombination by Int-wt. Reactions were performed using 32P-labeled attB or 32P-labeled attL and excess amounts of each of the 4 unlabeled att sites in the absence or presence of Gp44. Figure 7D shows representative gels for M117I and L121I, and the results for all mutants are given in Fig. S5 and summarized in Fig. 7E. F113Y and L121I exhibit att-site-specific, Gp44-regulated synapsis as SCs are observed only in attB × attP reactions in the absence of Gp44 and attL × attL or attR reactions in the presence of Gp44. L127V may fall into this class, but Gp44-activated attL-attL and attL-attR SCs were not apparent, although they may be obscured by the electrophoretic mobility shift generated by Gp44 binding (Fig. S5). The specificity of SC formation by these mutants correlates with their requirement for the CC motifs. In contrast, M117I exhibited promiscuous synapsis; SCs were generated with all combinations of att sites tested regardless of the presence of Gp44. Significantly, these included complexes involving attL and attR in the absence of Gp44, indicating that potential inhibitory CC motif interactions within a dimer bound to these sites are not preventing SC formation. G119Y formed significant amounts of attB-attP and all four complexes with attL in the presence of Gp44, albeit attL-attL and attL-attR were the most abundant of those formed with 32P-labeled attL (Fig. S5). The promiscuous nature of SC formation by M117I and, to a lesser extent, G119Y is consistent with their respective lack or reduced dependence on the CC motifs for synapsis (Fig. 7B).
We considered a model in which M117I is a tetramer in solution, thereby explaining its lack of att site specificity and CC independence of SC formation. The binding of a preassembled tetramer to one att site could then indiscriminately capture a second att site. Size exclusion chromatography was used to address the solution quaternary state of M117I. M117I is primarily a dimer in solution at concentrations well above those used in SC reactions, in both the presence (Fig. 7F) and the absence (not shown) of ethylene glycol. Under SC formation conditions in the presence of attP DNA, peaks consistent with the attP-bound dimer and tetrameric SCs were observed along with the free DNA (Fig. 7F). We conclude that the assembly pathway for SC formation by the promiscuous mutant M117I involves dimers binding to att sites followed by the DNA-bound dimers assembling into synaptic tetramers.
The SC-forming mutants were evaluated for recombination activity in intramolecular attP × attB reactions (Fig. 7G). These reactions were performed under standard recombination conditions since the addition of ethylene glycol inhibits recombination by Int-wt. All of the mutants exhibited reduced recombination efficiencies compared with the wt, with G119Y supporting about 83% and the others supporting between 31 and 46% of the wt reaction. No recombinants were detectable for the mutants deleted for their CC motifs except for G119Y, which gave a trace of intramolecular PB deletion products. Even M117I-ΔCC, which efficiently assembled stable SCs, did not generate detectable recombinants, including in reaction mixtures where ethylene glycol was added. The requirement of the CC motifs for recombination by M117I and G119Y suggests that the CC motifs may also function after initial synapsis, perhaps to facilitate complete isomerization to the active tetramer conformation (see Discussion).
Int-mediated DNA cleavage under SC-forming conditions was evaluated by quenching SC reactions performed with labeled DNA fragments or plasmid substrates using SDS or 0.1 M HCl. Only very small amounts of double-strand cleavages were observed for F113Y, M117I, G119Y, and L121I, and none was detected for L127V. The lack of efficient DNA cleavage contrasts with gain-of-function mutants at the analogous positions in the Hin invertase, which often form synaptic complexes containing the recombinase subunits covalently linked with the cleaved DNA ends (38, 39, 43).
DISCUSSION
In this work, we have explored mechanisms underlying the control of site-specific recombination reactions by the phage A118 serine integrase. We provide direct evidence that the identities of the four att recombination sites are determined solely by the spacing between the RD and ZD recognition sequences. The specificities of the reconstructed att sites confirm the model initially proposed by Van Duyne and coworkers based on their X-ray structure of the Int (L1)-attP complex and on point mutagenesis of the attP sequence (30, 35). The ZD is positioned on the same face (attP half-site) or opposite face (attB half-site) of the DNA helix relative to the RD (Fig. 1). The relative orientations of the ZD at each att half-site determine the trajectories of the CC motifs, which protrude from the ZD up to ∼100 Å from the DNA surface. The CC motif trajectories are further influenced by the binding of the RDF, as evidenced by the electrophoretic migrations of Int-att DNA complexes with and without Gp44 by native PAGE (17; this study). The electrophoretic migrations of the reconstructed att complexes recapitulate those of the native sites.
The tips of the CC motifs are proposed to interact at an early step in SC formation to juxtapose Int dimers bound to the correct att sites (30). This model predicts that the lengths of the CC arms would be important because they must extend beyond the surface of the catalytic domains to enable contacts between synapsing dimers (Fig. 1D). Indeed, we find that truncation of two helical turns from the middle of the CC motif generates a mutant that is nearly completely inactive for PB and LR recombination, but a construct that regains the length by addition of the same number of nonnative residues fully restores activity. Structural modeling of synaptic complexes suggests that the synapsing CC motifs must bend in order for the tips to interact with each other in a manner consistent with the X-ray structures of CC motif pairs and then for the dimers to isomerize into a tetramer to generate the chemically active synaptic complex (31). Such flexibility is consistent with the partial structures of the CC arms in the four complexes present in the L1-CTD attP crystal structure (30). Nevertheless, the extension of the length of the CC arms by two helical turns partially compromises PB and LR recombination. Strikingly, when the CC motifs are extended, Gp44 no longer efficiently inhibits PB recombination. The increased length of the flexible CC arms may enable CC tip interactions even when the RDF has altered their trajectories such that the wild-type length is insufficient.
A comprehensive alanine scan over 22 residues spanning the tip of the CC motif identified 14 residues as having significant roles in PB and/or LR recombination. The locations of these residues are consistent with the front-to-back interactions observed in the crystal structures and selective mutagenesis studies by Gupta et al. (31). Even though the mutagenesis data generally indicate that the contact surfaces are similar for PB and LR recombination, quantitative differences exhibited by a subset of the mutants imply that the molecular interactions between CC partners in the different recombination reactions are not identical. For example, K362A, F366A, L368A, Y374A, E378A, and D380A exhibit up to 10-fold-lower initial rates for PB than for LR recombination, whereas D367A is reduced 3-fold for LR recombination but unaltered for PB recombination. The differences in mutant activities may be correlated with differences in the interfaces of CC pairs revealed in crystal structures (Fig. 5A; see also Fig. S1 in the supplemental material). Although the current data are insufficient to make firm assignments, generally, the AB and EF pairs (PDB accession number 5U96) seem to better correlate with the attP × attB mutation data, and the CD and GH pairs better fit the attL × attR mutation data. Nonidentical CC interfaces within the two SC structures are consistent with the modeling of the attP-attB and attL-attR SCs, where different distortions to the CC helix regions are required in order to achieve pairing of the tips.
Initial CC-mediated pairing between subunits of synapsing dimers will localize the N-terminal ends of the E helices proximal to each other (Fig. 1D) in a configuration primed for remodeling into the active tetramer. Gupta et al. have measured pairing between CC units to occur with a dissociation constant (Kd) of ∼20 μM (31). Only dimers bound at attP and attB (or weakly between dimers bound to two attP sites for A118 Int) in either orientation and dimers bound at attL and attR (with the RDF) in the correct orientation for producing recombinants after DNA exchange are modeled to be able to form CC connections that juxtapose the N termini of the E helices from synapsing dimers. The role of the CC motifs in positioning the N termini of the E helices from synapsing dimers of serine integrases is analogous to the function of the Fis-bound enhancer for DNA inversion reactions or auxiliary resolvases and sometimes DNA bending proteins for deletion reactions by small SRs (46–48). Simultaneous opening of both dimers to expose their helix E surfaces is believed to trigger remodeling into the chemically active tetramer.
Forces controlling the dimer-tetramer transition during SC formation can be identified in gain-of-function mutants that lower the activation energy for remodeling by destabilizing the dimer and/or stabilizing the tetramer. Although SCs formed by A118 Int-wt are formed too transiently or are too unstable to be observed by native gel electrophoresis, amino acid substitutions at 5 positions within the E helix region allow SCs to be isolated. These gain-of-function mutants highlight the importance of the oligomerization E helices in controlling the dimer-tetramer conversion by serine integrases in a manner similar to that of small SRs. The residues that we have identified that control the dimer-tetramer conversion are predicted to be involved in (i) interactions between dimer subunits, (ii) interactions between helix E and the catalytic domain of the same subunit, or (iii) stabilization of the tetramer conformation by the mutant side chain. Our discussion that follows regarding the functional effects of the mutations is based on modeling of the A118 catalytic domain and helix E oligomerization region primarily in the dimer state (Fig. 7H) and detailed studies of analogous mutants in the small SRs (e.g., Hin, γδ/Tn3 resolvases, and Sin) (37–39, 42–44, 58).
A118 Int residues Phe113 and Leu127 engage in dimeric interactions and are modeled to transition through a complex set of interactions during the conversion of the dimer to the active tetramer (39, 44). Thus, the chemically similar mutant side chains present in A118 Int-F113Y and -L127V may partially destabilize the dimer and facilitate dimer-tetramer isomerization. Although sometimes observed with the other mutants, SCs generated by L127V consistently have lost one of the att site DNA fragments. The quaternary rearrangement may transition through a step that destabilizes the binding of one or possibly both synapsing DNA molecules.
The side chains of A118 Int-Met117 and -Leu121, where isoleucine substitutions promote stable SCs, are predicted to be positioned against the catalytic domain of the same subunit within the dimer (Fig. 7H). These are the positions of some of the strongest hyperactivating mutations in Hin (38). Modeling of the A118 Int dimer indicates that the aliphatic side chains are inserted into a cleft in the catalytic domain of the same polypeptide chain, similar to analogous residues in small SRs. During remodeling into the synaptic tetramer, the side chains rotate out of the cleft and are repositioned onto the surface of the interface that rotates during DNA exchange (38, 39, 44). Thus, changes in these residues are predicted to destabilize the dimer conformation. Although Hin mutants with changes at these positions efficiently form SCs that cleave DNA, they often fail to complete recombination because of ligation defects due to their position within the rotating interface of the tetramer. Substitutions could thus also destabilize the final active tetramer conformation.
M117I was the strongest of the A118 gain-of-function Int mutants that we isolated as it was the only one that efficiently formed SCs both without the CC motifs and without regard to the identities of the att sites. However, larger amounts of SCs containing attB and attP and, when Gp44 was present, attL and attR were formed over other att site combinations, indicating that the CC motifs still enhanced synapsis of the correct att sites (Fig. S5). The promiscuous nature of att site synapsis with the CC-independent mutant, in comparison to the stringent specificity of the other CC-dependent SC-forming mutants, provides strong support for the role of the CC motifs in controlling the att site specificity of synapsis.
We find it significant that M117I efficiently forms SCs with attL and attR in the absence of Gp44 because the CC motifs between dimeric subunits bound to these sites would be expected to interact and form a blocked complex. This result suggests that either the conformation of CC-bound dimers bound to attL and attR is competent for transitioning into a stable synaptic tetramer or the intradimer CC motif contacts are sufficiently dynamic to ineffectively block SC formation. The functional properties of the A118 Int-M117I mutation resemble those generated by the ϕC31 Int-E449K mutation, which is located at the base of the CC motif and believed to affect the trajectory of the CC motif. Like A118 Int-M117I, ϕC31 Int-E449K efficiently forms SCs without regard to the identity of att sites and the presence of the cognate RDF (32, 55).
G119Y is able to weakly form SCs in the absence of its CC motifs and exhibits promiscuous SC formation when Gp44 is present. The large side chain substitution is modeled to potentially clash with residues in the loop between β4 and β5 of the partner subunit and thereby destabilize the dimer, as is observed with the analogous Hin-H107Y mutant (39). A partially destabilized dimer would be consistent with the presence of small but reproducible amounts of monomer-bound att sites by G119Y, which are normally rarely observed (e.g., see Fig. S5 in the supplemental material). The G119Y substitution is likely to also stabilize the tetramer conformation in a manner similar to that of the Sin gain-of-function mutation Q117R. An X-ray structure of the Sin-Q117R tetramer shows the mutant arginines engaged in pi-stacking interactions that stabilize the synaptic tetramer (58). A similar model involving the stacking of mutant tyrosine side chains in the tetramer was proposed for the analogous Hin hyperactive mutant H107Y (39).
A difference between the A118 Int SC-forming mutants and most SC-forming mutants of the small SR family members is that the att sites within Int SCs are poorly or not at all (L127V) cleaved and covalently associated with the recombinase through a phosphoserine linkage. This could indicate that the Int proteins within these SCs are not fully isomerized to the chemically active conformation. It has been proposed that the remodeling of small SR dimers into the active tetramer may occur in two steps (39, 59). The first step involves the merging of the E helices from each dimer to generate a tetramer, whose conformation is represented by X-ray structures of the TP901 Int tetramer (PDB accession number 3BVP [49]) and one of the γδ resolvase tetramers (PDB accession number 2GM5 [60]). In this tetramer, the equivalents of A118 Int residues Met117 and Leu121 have rotated their side chains out of their catalytic domain pocket, but the chemically active catalytic site and flat rotating interface are not yet formed. The proteins within the stable SCs formed by the A118 helix E mutants may be in this intermediate, yet stable, conformation. A second conformational step that involves the rotation of the catalytic domain around the peptide backbone just prior to the start of helix E is required to generate the active tetramer competent for DNA cleavage and subunit rotation (39, 44). Consistent with this 2-step model, mutations at Val129 proximal to the start of helix E in ϕC31 Int can lead to the enhanced formation of SCs, but like the A118 mutants, these SCs exhibit low enzymatic activity (61). Similar phenotypes have been observed for mutations at the “hinge” residues Gly101 (and Asp/Glu102) in γδ/Tn3 resolvases and Ser99 in Hin (37, 39, 41, 44, 60).
Single-molecule studies by Fan and colleagues have provided evidence that ϕC31 Int forms short-lived abortive nonrecombinogenic SCs, in addition to productive SCs that generate recombinants (62). It is not clear if these complexes relate to the pathway to productive SCs or to the stable SCs identified by gel electrophoresis or SEC of the A118 Int helix E mutants. Whereas the effects of the E449K mutation, which is believed to affect the trajectory of the CC motifs, imply a role of the CC motifs in the formation of recombinogenic SCs (55), the abortive SCs form indiscriminately with regard to att site identity and the presence of the RDF (where tested).
In summary, the properties of the A118 helix E mutants demonstrate the interplay between the CC motifs and the E helices in controlling the recombination reaction. They provide strong evidence that the CC motifs function in, and are required for, the initial steps of att site synapsis where site identity and directionality (for attL-attR complexes) are evaluated as a function of the att site architecture. The M117I mutation in helix E overrides the requirement for the CC motifs for SC formation, and as a consequence, att site specificity for synapsis is lost. The CC motifs are still required for recombination by M117I, however, suggesting that the CC motifs also function at a step after initial synapsis, perhaps to complete tetramer remodeling to the active conformation. The importance of the particular helix E residues for synapsis in A118 Int suggests that principles learned from the small SRs regarding the control of synaptic tetramer formation are applicable to the LSRs.
MATERIALS AND METHODS
Plasmids and protein purification.
The descriptions of plasmids used in this work are summarized in Table S1 in the supplemental material. Oligonucleotides (IDT) specifying the reconstructed att sites (Fig. 2A) were annealed and ligated into the SmaI site of pUC18 to give pRJ3193 to pRJ3196, pRJ3231, and pRJ3232. Amino acid substitution mutations within the Int gene were generated in pRJ2184 (pET15b A118 int) or pRJ3259 (pRJ2184 intΔCC) by the QuikChange method using Q5 polymerase (New England BioLabs). Deletions or additions were generated by inverse PCR followed by ligation. Int and Gp44 proteins for purification were expressed in RJ3387 [Escherichia coli BL21(DE3) endA8::tet fis::kan-767]. N-terminally His-tagged Int proteins were expressed at 10°C from pRJ2184 and purified by chromatography through Ni-nitrilotriacetic acid (NTA) and heparin-Sepharose 6 Fast Flow resins as described previously (16). Intein-tagged Gp44 was expressed from pGV2710 (G. Van Duyne, University of Pennsylvania), purified using Ni-NTA and chitin resins as described previously (17), and then, following intein cleavage by 100 mM 2-mercaptoethanol, chromatographed through heparin-Sepharose 6 Fast Flow. Protein aliquots were stored in a solution containing 20 mM HEPES (pH 7.5), 0.3 M NaCl (Gp44) or 1 M NaCl (Int), 2 mM dithiothreitol (DTT), and 50% glycerol at −80°C.
Recombination reactions.
Intermolecular plasmid by linear DNA recombination reactions were performed under the conditions described previously (16, 17). Reaction mixtures contained 0.03 pmol plasmid DNA (pRJ2214 [attP], pRJ2215 [attB], pRJ2191 [attL], and pRJ2193 [attR]) and 0.09 pmol 100-bp att fragment DNA in a 20-μl solution containing 20 mM HEPES (pH 7.5), 100 mM KCl, 5 mM spermidine, 2.5 mM DTT, 5 mM MgCl2, 30 μg/ml bovine serum albumin (BSA), and 5% glycerol. Int (1 pmol dimer) or Int preincubated at 23°C with Gp44 (final amount, 4 pmol monomer) was added at 23°C, and the reaction mixture was incubated at 30°C. One-hundred-base-pair att fragments were generated by PCR using the above-mentioned plasmids or pRJ2986 and pRJ3244 as the templates and purified by PAGE. Reactions were terminated by SDS, and products were resolved on 1% agarose gels. Intramolecular reactions were performed similarly except that 0.02 pmol pRJ3244 (attB × attP) or pRJ2986 (attL × attR) was the substrate. After heat inactivation (65°C), the products were digested with NdeI to linearize each deletion circle product prior to agarose gel electrophoresis.
DNA binding assays: Int, Gp44, and synaptic complexes.
32P-labeled DNA fragments were generated by PCR after 5′ labeling of one or both primers by polynucleotide kinase and [γ-32P]ATP and purified by PAGE. DNA-binding reactions by Int and Int+Gp44, followed by electrophoresis on native polyacrylamide gels, were performed as described previously (16, 17).
Synaptic complex reactions were performed as follows. Int (0.5 to 1.0 pmol dimer) was added to a 20-μl reaction mix containing 20 mM HEPES (pH 7.5), 0.2 M KCl, 5 mM DTT, 0.1 mg/ml BSA, 25% ethylene glycol, 50 μg/ml sonicated salmon sperm DNA, 0.1 pmol labeled att DNA fragment, and 0.6 pmol unlabeled att DNA fragment. For reactions employing Gp44, Int and Gp44 (5-fold over the Int concentration) were preincubated in a solution containing 20 mM HEPES (pH 7.5), 0.3 M NaCl, 2 mM DTT, 0.1 mg/ml BSA, and 5% glycerol for 5 min at 23°C before adding them to the reaction mixture. Reaction mixtures were incubated for 10 min at 30°C and then applied to a 6% 39:1 acrylamide-bisacrylamide gel containing 10% glycerol in 0.5× Tris-borate-EDTA buffer at 4°C. DNA-binding gels were imaged using a Typhoon phosphorimager (Cytiva).
Size exclusion chromatography.
Int (20 μM) was buffer exchanged into 0.3 ml of a solution containing 20 mM HEPES (pH 7.5), 0.3 M KCl, 2 mM DTT, and 30% ethylene glycol containing 60 μM 60-bp attP duplex oligonucleotides, using a 0.5-ml Amicon Ultra filter unit (10,000 molecular weight [MW]; Millipore). After 15 min at 4°C, the reaction mixture was concentrated to about 0.1 ml with the Amicon Ultra filter unit and injected into a Superdex 200 16/60 column equilibrated in the same buffer except for 30% glycerol in place of ethylene glycol and chromatographed at a flow rate of 1 ml/min at 4°C.
Structural modeling.
Modeling of the A118 integrase dimer structure to att sites was performed with PyMOL (Schrödinger [http://www.pymol.org]) and Coot (63) using the structure of the phage L1 integrase CTD bound to the A118 attP half-site described previously by Van Duyne and coworkers (PDB accession number 4KIS [chains C, I, and J]) (30) and a model of the catalytic domain based on the γδ resolvase X-ray structure (PDB accession number 1GDT) (64). The L1 CTD is 97.5% identical to that of A118 integrase, and the helix E regions are identical in sequence. Intact CC motifs were appended from the structure under PDB accession number 5U96 (chain A) (31). DNA (helical twist of 34.2°) was generated by w3DNA (65).
Additional models of the A118 Int catalytic domain plus the helix E dimer structure were generated with Modeller (66) and Phyre2 (67). The A118 Int sequence was threaded onto a template of the structure under PDB accession number 3G13 (putative conjugative transposon recombinase dimer from Clostridium difficile [Modeller and Phyre2]) or 3GUV (resolvase family site-specific recombinase dimer from Streptococcus pneumoniae [Phyre2]). Alignment of A118 Int residues 1 to 145 by Clustal Omega gives 24% identity and 50% similarity with the structure under PDB accession number 3G13 and 30% identity and 59% similarity with the structure under accession number 3GUV. The relative positions, and conclusions based thereof, of the helix E gain-of-function mutants are essentially identical in the three models. However, the Phyre2 models appear off-register over the β4 region, which affects the catalytic domain core structure, due to the positioning of a 1-residue indel. Thus, we selected the PDB accession number 3G13-based model structure by Modeller for illustration. Structure figures were prepared using PyMOL.
ACKNOWLEDGMENT
This work was supported by NIH grant GM038509 (to R.C.J.).
Footnotes
Supplemental material is available online only.
Contributor Information
Reid C. Johnson, Email: rcjohnson@mednet.ucla.edu.
William W. Metcalf, University of Illinois at Urbana-Champaign
REFERENCES
- 1.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. 2015. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol 13:641–650. 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 2.Lewis JA, Hatfull GF. 2001. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res 29:2205–2216. 10.1093/nar/29.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fogg PC, Colloms S, Rosser S, Stark M, Smith MC. 2014. New applications for phage integrases. J Mol Biol 426:2703–2716. 10.1016/j.jmb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keravala A, Groth AC, Jarrahian S, Thyagarajan B, Hoyt JJ, Kirby PJ, Calos MP. 2006. A diversity of serine phage integrases mediate site-specific recombination in mammalian cells. Mol Genet Genomics 276:135–146. 10.1007/s00438-006-0129-5. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet J, Subsoontorn P, Endy D. 2012. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci U S A 109:8884–8889. 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colloms SD, Merrick CA, Olorunniji FJ, Stark WM, Smith MCM, Osbourn A, Keasling JD, Rosser SJ. 2014. Rapid metabolic pathway assembly and modification using serine integrase site-specific recombination. Nucleic Acids Res 42:e23. 10.1093/nar/gkt1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stark WM. 2017. Making serine integrases work for us. Curr Opin Microbiol 38:130–136. 10.1016/j.mib.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Siuti P, Yazbek J, Lu TK. 2013. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol 31:448–452. 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 9.Merrick CA, Zhao J, Rosser SJ. 2018. Serine integrases: advancing synthetic biology. ACS Synth Biol 7:299–310. 10.1021/acssynbio.7b00308. [DOI] [PubMed] [Google Scholar]
- 10.Chavez CL, Calos MP. 2011. Therapeutic applications of the phiC31 integrase system. Curr Gene Ther 11:375–381. 10.2174/156652311797415818. [DOI] [PubMed] [Google Scholar]
- 11.Grindley ND, Whiteson KL, Rice PA. 2006. Mechanisms of site-specific recombination. Annu Rev Biochem 75:567–605. 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 12.Smith MC, Thorpe HM. 2002. Diversity in the serine recombinases. Mol Microbiol 44:299–307. 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- 13.Smith MC. 2015. Phage-encoded serine integrases and other large serine recombinases. Microbiol Spectr 3:MDNA3-0059-2014. 10.1128/microbiolspec.MDNA3-0059-2014. [DOI] [PubMed] [Google Scholar]
- 14.Van Duyne GD, Rutherford K. 2013. Large serine recombinase domain structure and attachment site binding. Crit Rev Biochem Mol Biol 48:476–491. 10.3109/10409238.2013.831807. [DOI] [PubMed] [Google Scholar]
- 15.Loessner MJ, Inman RB, Lauer P, Calendar R. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol Microbiol 35:324–340. 10.1046/j.1365-2958.2000.01720.x. [DOI] [PubMed] [Google Scholar]
- 16.Mandali S, Dhar G, Avliyakulov NK, Haykinson MJ, Johnson RC. 2013. The site-specific integration reaction of Listeria phage A118 integrase, a serine recombinase. Mob DNA 4:2. 10.1186/1759-8753-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandali S, Gupta K, Dawson AR, Van Duyne GD, Johnson RC. 2017. Control of recombination directionality by the Listeria phage A118 protein Gp44 and the coiled-coil motif of its serine integrase. J Bacteriol 199:e00019-17. 10.1128/JB.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasechnek A, Rabinovich L, Stadnyuk O, Azulay G, Mioduser J, Argov T, Borovok I, Sigal N, Herskovits AA. 2020. Active lysogeny in Listeria monocytogenes is a bacteria-phage adaptive response in the mammalian environment. Cell Rep 32:107956. 10.1016/j.celrep.2020.107956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. 2012. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150:792–802. 10.1016/j.cell.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 20.Carrasco CD, Ramaswamy KS, Ramasubramanian TS, Golden JW. 1994. Anabaena xisF gene encodes a developmentally regulated site-specific recombinase. Genes Dev 8:74–83. 10.1101/gad.8.1.74. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel B, Losick R, Stragier P. 1990. The Bacillus subtilis gene for the development transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev 4:525–535. 10.1101/gad.4.4.525. [DOI] [PubMed] [Google Scholar]
- 22.Sato T, Samori Y, Kobayashi Y. 1990. The cisA cistron of Bacillus subtilis sporulation gene spoIVC encodes a protein homologous to a site-specific recombinase. J Bacteriol 172:1092–1098. 10.1128/JB.172.2.1092-1098.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bannam TL, Crellin PK, Rood JI. 1995. Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: the TnpX site-specific recombinase excises a circular transposon molecule. Mol Microbiol 16:535–551. 10.1111/j.1365-2958.1995.tb02417.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Smith MC, Mullany P. 2006. The conjugative transposon Tn5397 has a strong preference for integration into its Clostridium difficile target site. J Bacteriol 188:4871–4878. 10.1128/JB.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson CM, Grossman AD. 2015. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet 49:577–601. 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burrus V, Waldor MK. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol 155:376–386. 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Chen D, Peters BM, Li L, Li B, Xu Z, Shirliff ME. 2016. Staphylococcal chromosomal cassettes mec (SCCmec): a mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog 101:56–67. 10.1016/j.micpath.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 28.Misiura A, Pigli YZ, Boyle-Vavra S, Daum RS, Boocock MR, Rice PA. 2013. Roles of two large serine recombinases in mobilizing the methicillin-resistance cassette SCCmec. Mol Microbiol 88:1218–1229. 10.1111/mmi.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutherford K, Van Duyne GD. 2014. The ins and outs of serine integrase site-specific recombination. Curr Opin Struct Biol 24:125–131. 10.1016/j.sbi.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutherford K, Yuan P, Perry K, Sharp R, Van Duyne GD. 2013. Attachment site recognition and regulation of directionality by the serine integrases. Nucleic Acids Res 41:8341–8356. 10.1093/nar/gkt580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta K, Sharp R, Yuan JB, Li H, Van Duyne GD. 2017. Coiled-coil interactions mediate serine integrase directionality. Nucleic Acids Res 45:7339–7353. 10.1093/nar/gkx474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowley PA, Smith MCA, Younger E, Smith MCM. 2008. A motif in the C-terminal domain of phiC31 integrase controls the directionality of recombination. Nucleic Acids Res 36:3879–3891. 10.1093/nar/gkn269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fogg PCM, Younger E, Fernando BD, Khaleel T, Stark WM, Smith MCM. 2018. Recombination directionality factor gp3 binds phiC31 integrase via the zinc domain, potentially affecting the trajectory of the coiled-coil motif. Nucleic Acids Res 46:1308–1320. 10.1093/nar/gkx1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McEwan AR, Rowley PA, Smith MC. 2009. DNA binding and synapsis by the large C-terminal domain of phiC31 integrase. Nucleic Acids Res 37:4764–4773. 10.1093/nar/gkp485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Sharp R, Rutherford K, Gupta K, Van Duyne GD. 2018. Serine integrase attP binding and specificity. J Mol Biol 430:4401–4418. 10.1016/j.jmb.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnold PH, Blake DG, Grindley NDF, Boocock MR, Stark WM. 1999. Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. EMBO J 18:1407–1414. 10.1093/emboj/18.5.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burke ME, Arnold PH, He J, Wenwieser SV, Rowland SJ, Boocock MR, Stark WM. 2004. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol Microbiol 51:937–948. 10.1046/j.1365-2958.2003.03831.x. [DOI] [PubMed] [Google Scholar]
- 38.Chang Y, Johnson RC. 2015. Controlling tetramer formation, subunit rotation and DNA ligation during Hin-catalyzed DNA inversion. Nucleic Acids Res 43:6459–6472. 10.1093/nar/gkv565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heiss JK, Sanders ER, Johnson RC. 2011. Intrasubunit and intersubunit interactions controlling assembly of active synaptic complexes during Hin-catalyzed DNA recombination. J Mol Biol 411:744–764. 10.1016/j.jmb.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klippel A, Cloppenborg K, Kahmann R. 1988. Isolation and characterization of unusual Gin mutants. EMBO J 7:3983–3989. 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olorunniji FJ, He J, Wenwieser SV, Boocock MR, Stark WM. 2008. Synapsis and catalysis by activated Tn3 resolvase mutants. Nucleic Acids Res 36:7181–7191. 10.1093/nar/gkn885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowland SJ, Boocock MR, McPherson AL, Mouw KW, Rice PA, Stark WM. 2009. Regulatory mutations in Sin recombinase support a structure-based model of the synaptosome. Mol Microbiol 74:282–298. 10.1111/j.1365-2958.2009.06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders ER, Johnson RC. 2004. Stepwise dissection of the Hin-catalyzed recombination reaction from synapsis to resolution. J Mol Biol 340:753–766. 10.1016/j.jmb.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley ND, Steitz TA. 2005. Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs. Science 309:1210–1215. 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 45.Trejo CS, Rock RS, Stark WM, Boocock MR, Rice PA. 2018. Snapshots of a molecular swivel in action. Nucleic Acids Res 46:5286–5296. 10.1093/nar/gkx1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson RC. 2015. Site-specific DNA inversion by serine recombinases. Microbiol Spectr 3:MDNA3-0047-2014. 10.1128/microbiolspec.MDNA3-0047-2014. [DOI] [PubMed] [Google Scholar]
- 47.Rice PA. 2015. Serine resolvases. Microbiol Spectr 3:MDNA3-0045-2014. 10.1128/microbiolspec.MDNA3-0045-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stark WM. 2014. The serine recombinases. Microbiol Spectr 2:MDNA3-0046-2014. 10.1128/microbiolspec.MDNA3-0046-2014. [DOI] [PubMed] [Google Scholar]
- 49.Yuan P, Gupta K, Van Duyne GD. 2008. Tetrameric structure of a serine integrase catalytic domain. Structure 16:1275–1286. 10.1016/j.str.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh P, Kim AI, Hatfull GF. 2003. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol Cell 12:1101–1111. 10.1016/S1097-2765(03)00444-1. [DOI] [PubMed] [Google Scholar]
- 51.Smith MCA, Till R, Smith MCM. 2004. Switching the polarity of a bacteriophage integration system. Mol Microbiol 51:1719–1728. 10.1111/j.1365-2958.2003.03942.x. [DOI] [PubMed] [Google Scholar]
- 52.Stark WM, Grindley NDF, Hatfull GF, Boocock MR. 1991. Resolvase-catalysed reactions between res sites differing in the central dinucleotide of subsite I. EMBO J 10:3541–3548. 10.1002/j.1460-2075.1991.tb04918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moskowitz IP, Heichman KA, Johnson RC. 1991. Alignment of recombination sites in Hin-mediated site-specific DNA recombination. Genes Dev 5:1635–1645. 10.1101/gad.5.9.1635. [DOI] [PubMed] [Google Scholar]
- 54.Ghosh P, Bibb LA, Hatfull GF. 2008. Two-step site selection for serine-integrase-mediated excision: DNA-directed integrase conformation and central dinucleotide proofreading. Proc Natl Acad Sci U S A 105:3238–3243. 10.1073/pnas.0711649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan HF, Su BY, Ma CH, Rowley PA, Jayaram M. 2020. A bipartite thermodynamic-kinetic contribution by an activating mutation to RDF-independent excision by a phage serine integrase. Nucleic Acids Res 48:6413–6430. 10.1093/nar/gkaa401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith MCA, Till R, Brady K, Soultanas P, Thorpe H, Smith MCM. 2004. Synapsis and DNA cleavage in phiC31 integrase-mediated site-specific recombination. Nucleic Acids Res 32:2607–2617. 10.1093/nar/gkh538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olorunniji FJ, McPherson AL, Pavlou HJ, McIlwraith MJ, Brazier JA, Cosstick R, Stark WM. 2015. Nicked-site substrates for a serine recombinase reveal enzyme-DNA communications and an essential tethering role of covalent enzyme-DNA linkages. Nucleic Acids Res 43:6134–6143. 10.1093/nar/gkv521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keenholtz RA, Rowland SJ, Boocock MR, Stark WM, Rice PA. 2011. Structural basis for catalytic activation of a serine recombinase. Structure 19:799–809. 10.1016/j.str.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dhar G, McLean MM, Heiss JK, Johnson RC. 2009. The Hin recombinase assembles a tetrameric protein swivel that exchanges DNA strands. Nucleic Acids Res 37:4743–4756. 10.1093/nar/gkp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamtekar S, Ho RS, Cocco MJ, Li W, Wenwieser SV, Boocock MR, Grindley NDF, Steitz TA. 2006. Implications of structures of synaptic tetramers of gamma delta resolvase for the mechanism of recombination. Proc Natl Acad Sci U S A 103:10642–10647. 10.1073/pnas.0604062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rowley PA, Smith MC. 2008. Role of the N-terminal domain of phiC31 integrase in attB-attP synapsis. J Bacteriol 190:6918–6921. 10.1128/JB.00612-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan HF, Hsieh TS, Ma CH, Jayaram M. 2016. Single-molecule analysis of varphiC31 integrase-mediated site-specific recombination by tethered particle motion. Nucleic Acids Res 44:10804–10823. 10.1093/nar/gkw861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Yang W, Steitz TA. 1995. Crystal structure of the site-specific recombinase gammadelta resolvase complexed with a 34 bp cleavage site. Cell 82:193–207. 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 65.Li S, Olson WK, Lu XJ. 2019. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res 47:W26–W34. 10.1093/nar/gkz394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webb B, Sali A. 2016. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 54:5.6.1–5.6.37. 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 Web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5 and Table S1. Download JB00703-20_Supp_1_seq9.pdf, PDF file, 2.2 MB (2.2MB, pdf)