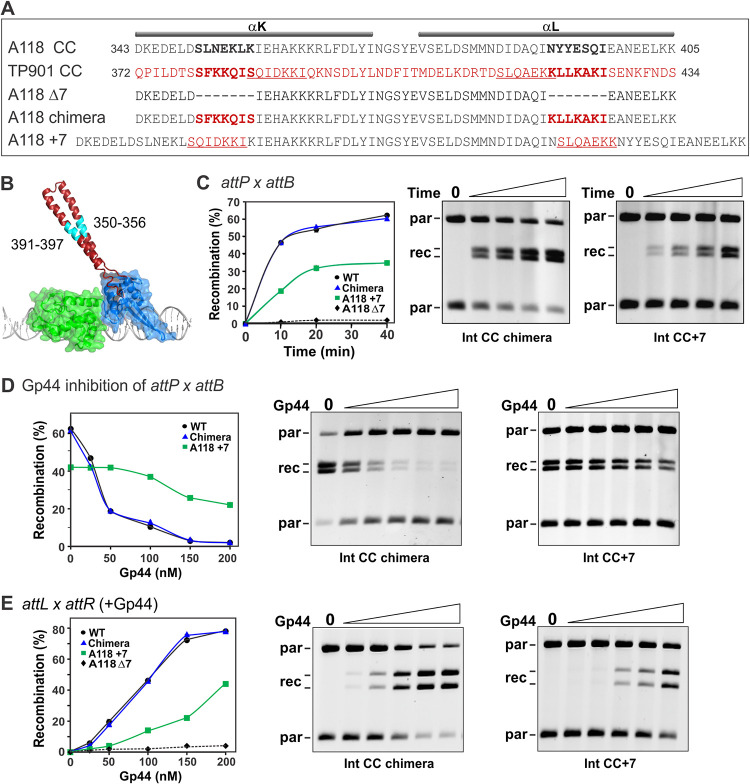
FIG 4.
The length of the CC motif is important for activity. (A) Alignment of the amino acid residue sequences of the CC segments from the A118 and TP901 integrases. Below are the sequences of the CC length mutants. (B) Structure of the L1 integrase RD (green) and ZD (blue) rendered as a transparent surface. The CC motif (PDB accession number 4KIS for chain C with the tip appended from PDB accession number 5U96 for chain A) is highlighted in red; cyan denotes the segment deleted in A118 CCΔ7 or replaced with TP901 residues (A118 chimera). (C) attP × attB deletion reactions by the wt and CC mutant integrases. On the left are plots of representative reaction time courses, and on the right are gel images of reactions by the Int CC chimera and Int CC+7 mutants. The DNA was digested with NdeI, which cleaves the parental (par) plasmid twice and linearizes the product circles, prior to agarose gel electrophoresis (see Fig. S4 in the supplemental material). (D) Gp44 inhibition of attP × attB deletion reactions by wt and mutant integrases. Reactions were performed as described above for panel C for 20 min in the presence of 0, 25, 50, 100, 150, and 200 nM Gp44, and example data are shown. (E) Gp44-activated attL × attR deletion reactions by the wt and CC mutant integrases. Reactions were performed as described above for panel D.