FIGURE 2.
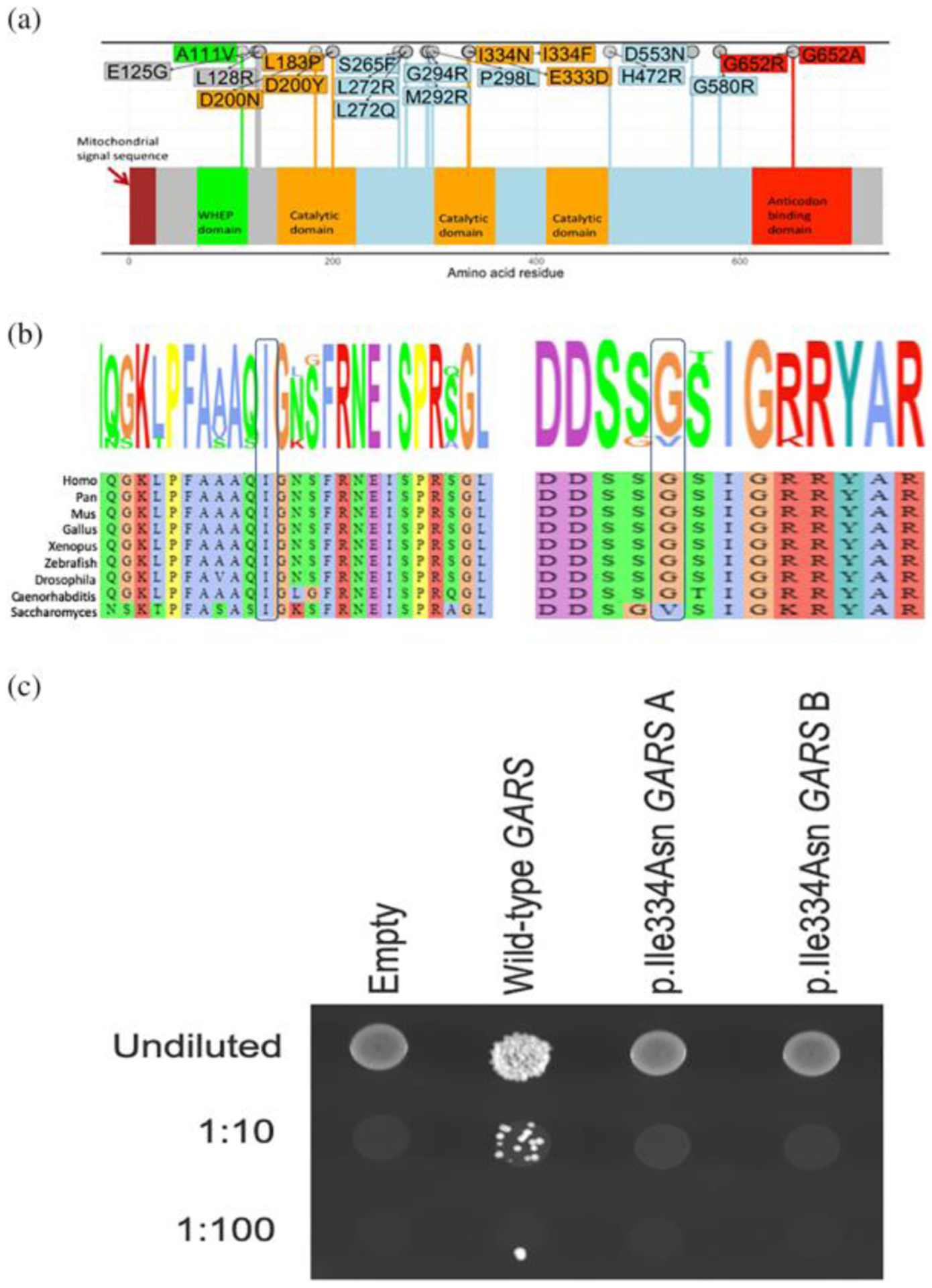
(a) The mutation spectrum of GARS. Missense mutations causing CMT2D/ dSMA are shown on the GARS primary structure. The domains are colored as indicated. (b) Conservation of the p. Ile334 and the p.Gly652 residues. (c) The p.Ile334Asn GARS variant causes a loss-of-function effect in yeast complementation assays. Yeast lacking endogenous GRS1 (the yeast ortholog of GARS) were transformed with vectors containing wild-type GARS ΔMTSΔWHEP or p.Ile334Asn GARS ΔMTSΔWHEP, or a vector with no GARS insert (“Empty”). Resulting cultures were plated undiluted or diluted (1:10 or 1:100) on media containing 5-FOA or media lacking leucine and uracil (−leu-ura) and grown at 30°C. Two independently generated mutant constructs (“a” and “b”) were tested
