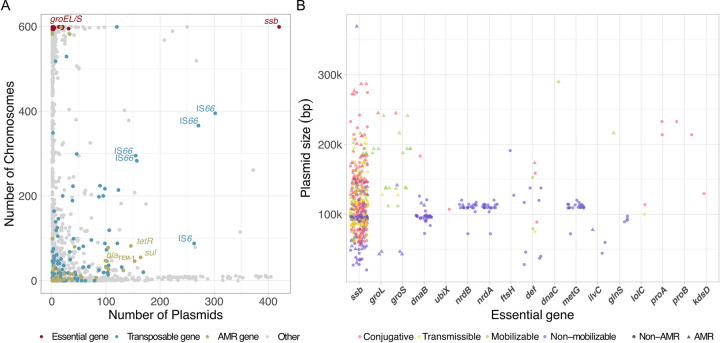
Fig 1. The distribution of plasmid-encoded essential genes on plasmids and chromosomes.
A, The distribution of Escherichia protein families on plasmids in chromosomes (in grey). Essential genes (red) are found in most of the chromosomes and rarely on plasmids, with ssb as an exception. Transposable elements (blue) are frequent on both chromosomes and plasmids. IS66 is the most widely spread gene in Escherichia strains. IS66 was split into several protein families in our analysis, which are depicted by multiple data points. Antibiotic resistance (AMR) genes (yellow) are presented for comparison. The AMR genes can be roughly divided into two groups: the first group aligns along the y-axis hence it is more frequently found on chromosomes; those chromosome-encoded AMR genes are typically related to persistence and resilience functions. The second group aligns along the x-axis hence it is more frequently found on plasmids; those plasmid-encoded genes are typically related to antibiotics resistance functions (Wein et al. 2020). B, Distribution of plasmid size, mobility group and AMR for plasmids encoding an essential gene. Most of the ssb-coding plasmids are conjugative (red) while the majority of groEL/S coding plasmids are mobilizable (green). The remaining essential genes are encoded on plasmids that are often non-mobilizable (and non-AMR) (purple).