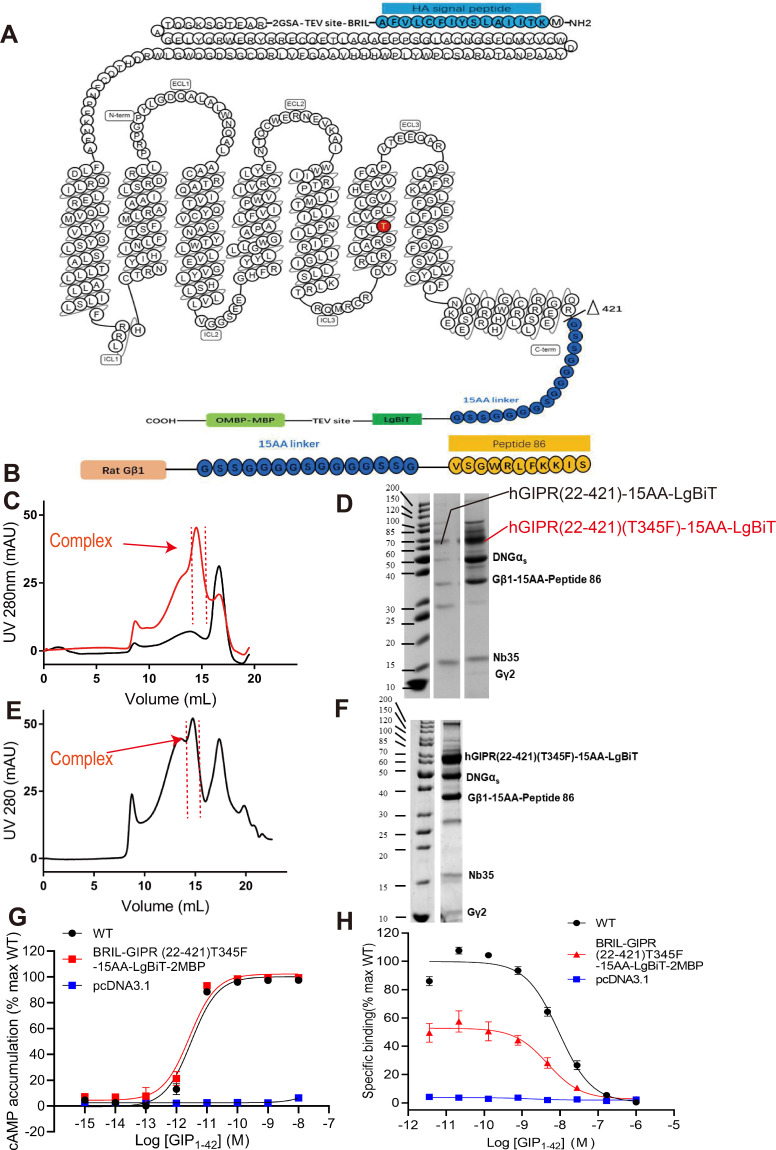
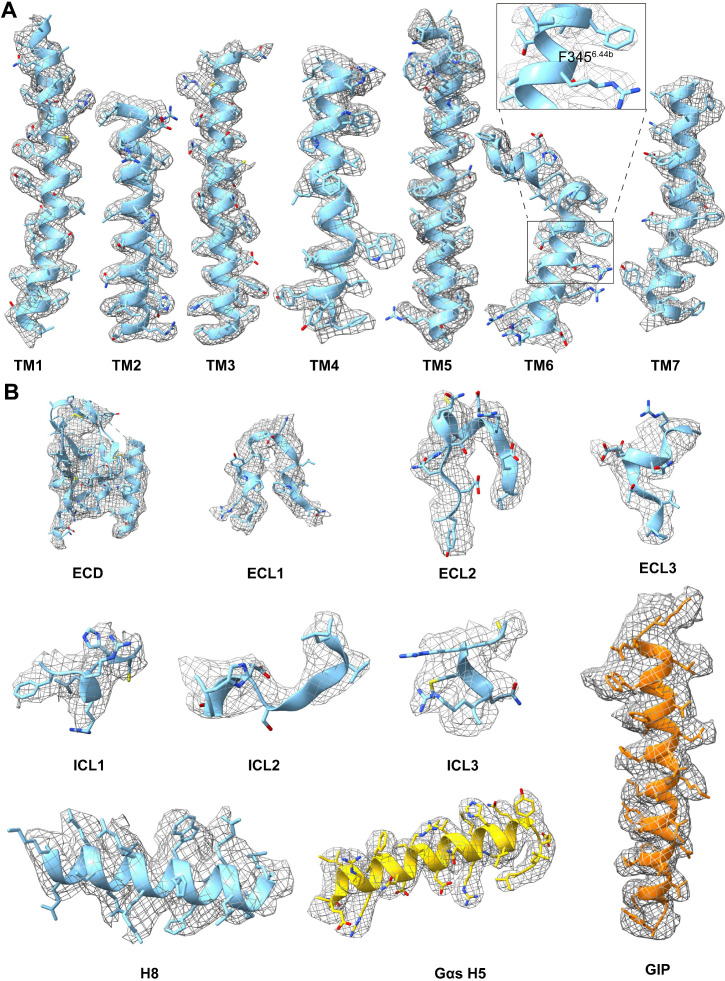
Figure 1. Cryo-EM structure of the GIP1-42–GIPR–Gs complex.
(A) Cut-through view of the cryo-EM density map that illustrates the GIP1-42–GIPR–Gs complex and the disc-shaped micelle. The unsharpened cryo-EM density map at the 0.07 threshold shown as light gray surface indicates a micelle diameter of 11 nm. The colored cryo-EM density map is shown at the 0.16 threshold. (B) Model of the complex as a cartoon, with GIP1-42 as helix in orange. The receptor is shown in light sky blue, Gαs in yellow, Gβ subunit in cyan, Gγ subunit in navy blue, and Nb35 in gray.