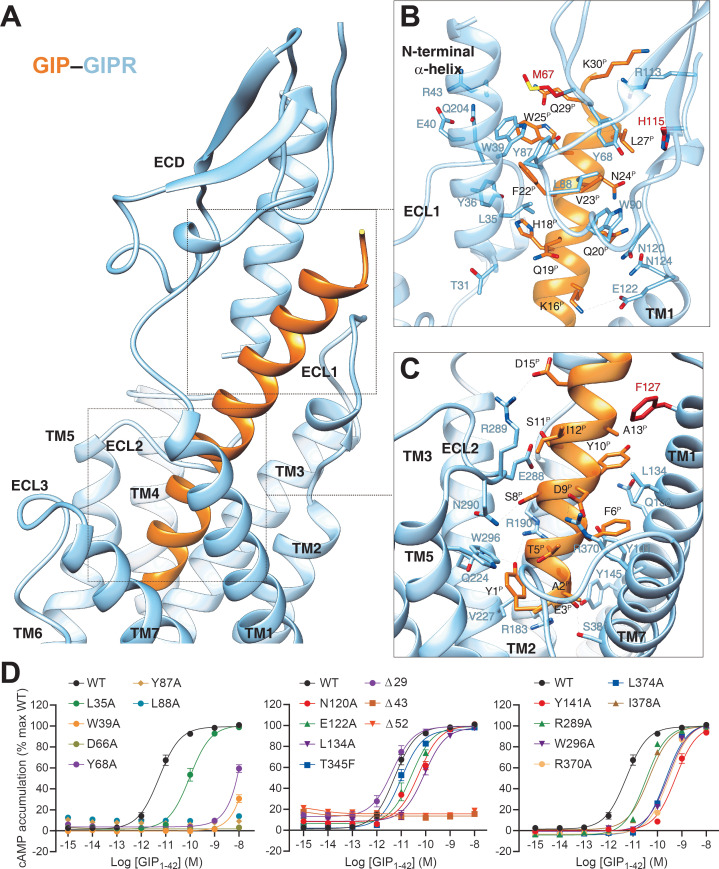
Figure 2. Molecular recognition of GIP by GIPR.
(A) The binding mode of GIP (orange) with GIPR (light sky blue), showing that the N-terminal half of GIP penetrates into a pocket formed by all TM helices except TM4, ECL2, and ECL3, whereas the C-terminal half is recognized by ECD, ECL1, and TM1. (B, C) Close-up views of the interactions between GIP and GIPR. The residues and side chains that could not be modelled in the ECD are colored in red. (D) Signaling profiles of GIPR mutants. cAMP accumulation in wild-type (WT) and single-point mutated GIPR expressing in HEK 293T cells. Signals were normalized to the maximum response of the WT and dose–response curves were analyzed using a three-parameter logistic equation. All data were generated and graphed as means ± S.E.M. of at least three independent experiments, conducted in quadruplicate. Δ, truncated residues.